American Museum Novitates
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Seasonal Shedding of Coronaviruses in Straw-Colored Fruit Bats at Urban Roosts in Africa
Seasonal Shedding of Coronaviruses in Straw-colored Fruit Bats at Urban Roosts in Africa The adaptation of bats (order Chiroptera) to use and For these reasons, we assessed the seasonality of occupy human dwellings across the planet has created coronavirus (CoV) shedding by the straw-colored intensive bat-human interfaces. Because bats provide fruit bat (Eidolon helvum) by passively collecting 97 important ecosystem services and also host and shed fecal samples on a monthly basis during a entire year zoonotic viruses, these interfaces represent a double in two urban colonies: Accra, Ghana (West Africa) challenge: i) the conservation of bats and their services and Morogoro, Tanzania (East Africa; Fig 1). Sampling and ii) the prevention of viral spillover. collection was conducted under the same trees during Many species of bats have evolved a seasonal life the study period. This species of fruit bat shows a single history that has resulted in the development of specific birth pulse during the year, its colonies show spectacular reproductive and foraging activities during distinctive periodical changes in size, and similarly to other tree- periods of the year. For example, many species mate, roosting megabats, several roosts are located in busy give birth, and nurse during particular and predictable urban centers across sub-Saharan Africa. Moreover, times of the year. Moreover, bat migration can produce we concomitantly collected data on the roost sizes predictable variations in colony sizes during a typical and precipitation levels over time, and we established year, from a complete absence of bats to the aggregation the reproductive periods through the year (birth of millions of individuals depending on the season. -

A Recent Bat Survey Reveals Bukit Barisan Selatan Landscape As A
A Recent Bat Survey Reveals Bukit Barisan Selatan Landscape as a Chiropteran Diversity Hotspot in Sumatra Author(s): Joe Chun-Chia Huang, Elly Lestari Jazdzyk, Meyner Nusalawo, Ibnu Maryanto, Maharadatunkamsi, Sigit Wiantoro, and Tigga Kingston Source: Acta Chiropterologica, 16(2):413-449. Published By: Museum and Institute of Zoology, Polish Academy of Sciences DOI: http://dx.doi.org/10.3161/150811014X687369 URL: http://www.bioone.org/doi/full/10.3161/150811014X687369 BioOne (www.bioone.org) is a nonprofit, online aggregation of core research in the biological, ecological, and environmental sciences. BioOne provides a sustainable online platform for over 170 journals and books published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Web site, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/page/terms_of_use. Usage of BioOne content is strictly limited to personal, educational, and non-commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. Acta Chiropterologica, 16(2): 413–449, 2014 PL ISSN 1508-1109 © Museum and Institute of Zoology PAS doi: 10.3161/150811014X687369 A recent -

Chiroptera: Pteropodidae)
Chapter 6 Phylogenetic Relationships of Harpyionycterine Megabats (Chiroptera: Pteropodidae) NORBERTO P. GIANNINI1,2, FRANCISCA CUNHA ALMEIDA1,3, AND NANCY B. SIMMONS1 ABSTRACT After almost 70 years of stability following publication of Andersen’s (1912) monograph on the group, the systematics of megachiropteran bats (Chiroptera: Pteropodidae) was thrown into flux with the advent of molecular phylogenetics in the 1980s—a state where it has remained ever since. One particularly problematic group has been the Austromalayan Harpyionycterinae, currently thought to include Dobsonia and Harpyionycteris, and probably also Aproteles.Inthis contribution we revisit the systematics of harpyionycterines. We examine historical hypotheses of relationships including the suggestion by O. Thomas (1896) that the rousettine Boneia bidens may be related to Harpyionycteris, and report the results of a series of phylogenetic analyses based on new as well as previously published sequence data from the genes RAG1, RAG2, vWF, c-mos, cytb, 12S, tVal, 16S,andND2. Despite a striking lack of morphological synapomorphies, results of our combined analyses indicate that Boneia groups with Aproteles, Dobsonia, and Harpyionycteris in a well-supported, expanded Harpyionycterinae. While monophyly of this group is well supported, topological changes within this clade across analyses of different data partitions indicate conflicting phylogenetic signals in the mitochondrial partition. The position of the harpyionycterine clade within the megachiropteran tree remains somewhat uncertain. Nevertheless, biogeographic patterns (vicariance-dispersal events) within Harpyionycterinae appear clear and can be directly linked to major biogeographic boundaries of the Austromalayan region. The new phylogeny of Harpionycterinae also provides a new framework for interpreting aspects of dental evolution in pteropodids (e.g., reduction in the incisor dentition) and allows prediction of roosting habits for Harpyionycteris, whose habits are unknown. -
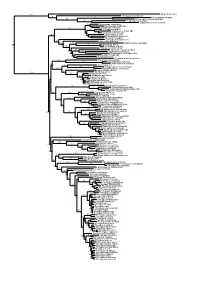
Figs1 ML Tree.Pdf
100 Megaderma lyra Rhinopoma hardwickei 71 100 Rhinolophus creaghi 100 Rhinolophus ferrumequinum 100 Hipposideros armiger Hipposideros commersoni 99 Megaerops ecaudatus 85 Megaerops niphanae 100 Megaerops kusnotoi 100 Cynopterus sphinx 98 Cynopterus horsfieldii 69 Cynopterus brachyotis 94 50 Ptenochirus minor 86 Ptenochirus wetmorei Ptenochirus jagori Dyacopterus spadiceus 99 Sphaerias blanfordi 99 97 Balionycteris maculata 100 Aethalops alecto 99 Aethalops aequalis Thoopterus nigrescens 97 Alionycteris paucidentata 33 99 Haplonycteris fischeri 29 Otopteropus cartilagonodus Latidens salimalii 43 88 Penthetor lucasi Chironax melanocephalus 90 Syconycteris australis 100 Macroglossus minimus 34 Macroglossus sobrinus 92 Boneia bidens 100 Harpyionycteris whiteheadi 69 Harpyionycteris celebensis Aproteles bulmerae 51 Dobsonia minor 100 100 80 Dobsonia inermis Dobsonia praedatrix 99 96 14 Dobsonia viridis Dobsonia peronii 47 Dobsonia pannietensis 56 Dobsonia moluccensis 29 Dobsonia anderseni 100 Scotonycteris zenkeri 100 Casinycteris ophiodon 87 Casinycteris campomaanensis Casinycteris argynnis 99 100 Eonycteris spelaea 100 Eonycteris major Eonycteris robusta 100 100 Rousettus amplexicaudatus 94 Rousettus spinalatus 99 Rousettus leschenaultii 100 Rousettus aegyptiacus 77 Rousettus madagascariensis 87 Rousettus obliviosus Stenonycteris lanosus 100 Megaloglossus woermanni 100 91 Megaloglossus azagnyi 22 Myonycteris angolensis 100 87 Myonycteris torquata 61 Myonycteris brachycephala 33 41 Myonycteris leptodon Myonycteris relicta 68 Plerotes anchietae -

Ecological Assessments in the B+WISER Sites
Ecological Assessments in the B+WISER Sites (Northern Sierra Madre Natural Park, Upper Marikina-Kaliwa Forest Reserve, Bago River Watershed and Forest Reserve, Naujan Lake National Park and Subwatersheds, Mt. Kitanglad Range Natural Park and Mt. Apo Natural Park) Philippines Biodiversity & Watersheds Improved for Stronger Economy & Ecosystem Resilience (B+WISER) 23 March 2015 This publication was produced for review by the United States Agency for International Development. It was prepared by Chemonics International Inc. The Biodiversity and Watersheds Improved for Stronger Economy and Ecosystem Resilience Program is funded by the USAID, Contract No. AID-492-C-13-00002 and implemented by Chemonics International in association with: Fauna and Flora International (FFI) Haribon Foundation World Agroforestry Center (ICRAF) The author’s views expressed in this publication do not necessarily reflect the views of the United States Agency for International Development or the United States Government. Ecological Assessments in the B+WISER Sites Philippines Biodiversity and Watersheds Improved for Stronger Economy and Ecosystem Resilience (B+WISER) Program Implemented with: Department of Environment and Natural Resources Other National Government Agencies Local Government Units and Agencies Supported by: United States Agency for International Development Contract No.: AID-492-C-13-00002 Managed by: Chemonics International Inc. in partnership with Fauna and Flora International (FFI) Haribon Foundation World Agroforestry Center (ICRAF) 23 March -

Priority-Setting for Philippine Bats Using Practical Approach to Guide Effective Species Conservation and Policy-Making in the Anthropocene
Published by Associazione Teriologica Italiana Volume 30 (1): 74–83, 2019 Hystrix, the Italian Journal of Mammalogy Available online at: http://www.italian-journal-of-mammalogy.it doi:10.4404/hystrix–00172-2019 Research Article Priority-setting for Philippine bats using practical approach to guide effective species conservation and policy-making in the Anthropocene Krizler Cejuela Tanalgo1,2,3,4,∗, Alice Catherine Hughes1,3 1Landscape Ecology Group, Centre for Integrative Conservation, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Menglun, Mengla, Yunnan Province 666303, People’s Republic of China 2International College, University of Chinese Academy of Sciences, Beijing 100049, People’s Republic of China 3Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences, Menglun, Mengla, Yunnan Province 666303, People’s Republic of China 4Department of Biological Sciences, College of Arts and Sciences, University of Southern Mindanao, Kabacan 9407, North Cotabato, the Republic of the Philippines Keywords: Abstract asian tropics endemism National level approaches to the development and implementation of effective conservation policy forest loss and practice are often challenged by limited capacity and resources. Developing relevant and hunting achievable priorities at the national level is a crucial step for effective conservation. The Philip- islands pine archipelago includes over 7000 islands and is one of only two countries considered both a oil palm global biodiversity hotspot and a megadiversity country. Yet, few studies have conducted over- arching synthesis for threats and conservation priorities of any species group. As bats make up a Article history: significant proportion of mammalian diversity in the Philippines and fulfil vital roles to maintain Received: 01/19/2019 ecosystem health and services we focus on assessing the threats and priorities to their conserva- Accepted: 24/06/2019 tion across the Philippines. -

Bat Count 2003
BAT COUNT 2003 Working to promote the long term, sustainable conservation of globally threatened flying foxes in the Philippines, by developing baseline population information, increasing public awareness, and training students and protected area managers in field monitoring techniques. 1 A Terminal Report Submitted by Tammy Mildenstein1, Apolinario B. Cariño2, and Samuel Stier1 1Fish and Wildlife Biology, University of Montana, USA 2Silliman University and Mt. Talinis – Twin Lakes Federation of People’s Organizations, Diputado Extension, Sibulan, Negros Oriental, Philippines Photo by: Juan Pablo Moreiras 2 EXECUTIVE SUMMARY Large flying foxes in insular Southeast Asia are the most threatened of the Old World fruit bats due to deforestation, unregulated hunting, and little conservation commitment from local governments. Despite the fact they are globally endangered and play essential ecological roles in forest regeneration as seed dispersers and pollinators, there have been only a few studies on these bats that provide information useful to their conservation management. Our project aims to promote the conservation of large flying foxes in the Philippines by providing protected area managers with the training and the baseline information necessary to design and implement a long-term management plan for flying foxes. We focused our efforts on the globally endangered Philippine endemics, Acerodon jubatus and Acerodon leucotis, and the bats that commonly roost with them, Pteropus hypomelanus, P. vampyrus lanensis, and P. pumilus which are thought to be declining in the Philippines. Local participation is an integral part of our project. We conducted the first national training workshop on flying fox population counts and conservation at the Subic Bay area. -

ANTIQUITY 2008 (In Press) an Unexpected, Stripe-Faced Flying Fox in Ice Age Rock Art of Australia's Kimberley. “Jack” Pett
ANTIQUITY 2008 (in press) An Unexpected, Stripe-faced Flying Fox in Ice Age Rock Art of Australia’s Kimberley. “Jack” Pettigrew, Marilyn Nugent, Anscar McPhee, Josh Wallman Bradshaw rock art of northern Australia enjoys continuing controversy concerning what community painted them and how to interpret the images (Roy 2002). The keen observation and accurate depictions of the natural world shown here, as well as the extraordinary longevity of the stains used, all tend to support the side of the controversy that posits a distinct cultural entity. We describe a painting unmistakably depicting flying foxes with features not found in bats presently found in Australia. Thermoluminescence dating of wasp nest overlying the art suggests an ice age migration to Australia, either of the bats or of the artists who painted them, a more likely scenario biologically than younger dates. The bat depictions were found on a sandstone wall protected by overhangs, near Kalumburu (14.30 °S; 126.64 °E), amongst other walls showing characteristic Bradshaw art (Walsh 2002). The depiction shows eight roosting megabats (flying foxes, Family Pteropodidae, sub-Order Megachiroptera) hanging from a slender branch, or more likely, a vine (Figs 1,2). Each bat has a distinctive white facial stripe and pale belly (Fig. 3). Figure1. White-striped flying foxes depicted in Bradshaw rock art at Kalumburu, in the Kimberley of Western Australia. Figure 2. Extension, to the left, of the same depicted group of shown in Fig. 1, with which there is some overlap. Dating: The indelible inks used in Bradshaw art penetrate more than a millimetre into the sandstone but have resisted all attempts so far to date them directly, with a number of different estimates of their age (Michaelson and Ebersole 2000). -

Conventional Wisdom on Roosting Behaviour of Australian
Conventional wisdom on roosting behaviour of Australian flying foxes - a critical review, and evaluation using new data Tamika Lunn1, Peggy Eby2, Remy Brooks1, Hamish McCallum1, Raina Plowright3, Maureen Kessler4, and Alison Peel1 1Griffith University 2University of New South Wales 3Montana State University 4Montana State University System April 12, 2021 Abstract 1. Fruit bats (Family: Pteropodidae) are animals of great ecological and economic importance, yet their populations are threatened by ongoing habitat loss and human persecution. A lack of ecological knowledge for the vast majority of Pteropodid bat species presents additional challenges for their conservation and management. 2. In Australia, populations of flying-fox species (Genus: Pteropus) are declining and management approaches are highly contentious. Australian flying-fox roosts are exposed to management regimes involving habitat modification, either through human-wildlife conflict management policies, or vegetation restoration programs. Details on the fine-scale roosting ecology of flying-foxes are not sufficiently known to provide evidence-based guidance for these regimes and the impact on flying-foxes of these habitat modifications is poorly understood. 3. We seek to identify and test commonly held understandings about the roosting ecology of Australian flying-foxes to inform practical recommendations and guide and refine management practices at flying-fox roosts. 4. We identify 31 statements relevant to understanding of flying-fox roosting structure, and synthesise these in the context of existing literature. We then contribute contemporary data on the fine-scale roosting structure of flying-fox species in south-eastern Queensland and north- eastern New South Wales, presenting a 13-month dataset from 2,522 spatially referenced roost trees across eight sites. -

Final Report on the Project
BP Conservation Programme (CLP) 2005 PROJECT NO. 101405 - BRONZE AWARD WINNER ECOLOGY, DISTRIBUTION, STATUS AND PROTECTION OF THREE CONGOLESE FRUIT BATS FINAL REPORT Patrick KIPALU Team Leader Observatoire Congolais pour la Protection de l’Environnement OCPE – ong Kinshasa – Democratic Republic of the Congo E-mail: [email protected] APRIL 2009 1 Table of Content Acknowledgements…………………………………………………………………. p3 I. Project Summary……………………………………………………………….. p4 II. Introduction…………………………………………………………………… p4-p7 III. Materials and Methods ……………………………………………………….. p7-p10 IV. Results per Study Site…………………………………………………………. p10-p15 1. Pointe-Noire ………………………………………………………….. p10-p12 2. Mayumbe Forest /Luki Reserve……………………………………….. p12-p13 3. Zongo Forest…………………………………………………………... p14 4. Mbanza-Ngungu ………………………………………………………. P15 V. General Results ………………………………………………………………p15-p16 VI. Discussions……………………………………………………………………p17-18 VII. Conclusion and Recommendations……………………………………….p 18-p19 VIII. Bibliography………………………………………………………………p20-p21 Acknowledgements 2 The OCPE (Observatoire Congolais pour la Protection de l’Environnement) project team would like to start by expressing our gratefulness and saying thank you to the BP Conservation Program, which has funded the execution of this project. The OCPE also thanks the Van Tienhoven Foundation which provided a further financial support. Without these organisations, execution of the project would not have been possible. We would like to thank specially the BPCP “dream team”: Marianne D. Carter, Robyn Dalzen and our regretted Kate Stoke for their time, advices, expertise and care, which helped us to complete this work, Our special gratitude goes to Dr. Wim Bergmans, who was the hero behind the scene from the conception to the execution of the research work. Without his expertise, advices and network it would had been difficult for the project team to produce any result from this project. -

NHBSS 042 1O Robinson Obs
Notes NAT. NAT. HIST. B 肌 L. SIAM Soc. 42: 117-120 , 1994 Observation Observation on the Wildlife Trade at the Daily Market in Chiang Kh an , Northeast Thailand In In a recent study into the wildlife trade between Lao PDR. and Thailand S即 KOSAMAT 組 A et al. (1 992) found 伽 tit was a major 伽 eat to wildlife resources in Lao. Th ey surveyed 15 locations along 白e 百凶 Lao border ,including Chiang Kh an. Only a single single visit was made to the market in Chiang Kh佃, on 8 April 1991 , and no wildlife products products were found. τbe border town of Chi 佃 g Kh an , Loei Pr ovince in northeast 百lailand is situated on the the banks of the Mekong river which sep 釘 ates Lao PD R. and 官 lailand. In Chiang Kh an there there is a small market held twice each day. Th e market ,situated in the centre of town , sells sells a wide r佃 ge of products from f記sh fruit ,vegetables , meat and dried fish to clothes and and household items. A moming market begins around 0300 h and continues until about 0800 h. The evening market st 紅白紙 1500 h and continues until about 1830 h. During During the period November 1992 to November 1993 visits were made to Chiang Kh佃 eve 凶ng market , to record the wildlife offered for sale. Only the presence of mam- mals ,reptiles and birds was recorded. Although frogs ,insects ,fish , crabs and 加 t eggs , as as well as meat 仕om domestic animals , were sold ,no record of this was made. -

Species-Edition-Melanesian-Geo.Pdf
Nature Melanesian www.melanesiangeo.com Geo Tranquility 6 14 18 24 34 66 72 74 82 6 Herping the final frontier 42 Seahabitats and dugongs in the Lau Lagoon 10 Community-based response to protecting biodiversity in East 46 Herping the sunset islands Kwaio, Solomon Islands 50 Freshwater secrets Ocean 14 Leatherback turtle community monitoring 54 Freshwater hidden treasures 18 Monkey-faced bats and flying foxes 58 Choiseul Island: A biogeographic in the Western Solomon Islands stepping-stone for reptiles and amphibians of the Solomon Islands 22 The diversity and resilience of flying foxes to logging 64 Conservation Development 24 Feasibility studies for conserving 66 Chasing clouds Santa Cruz Ground-dove 72 Tetepare’s turtle rodeo and their 26 Network Building: Building a conservation effort network to meet local and national development aspirations in 74 Secrets of Tetepare Culture Western Province 76 Understanding plant & kastom 28 Local rangers undergo legal knowledge on Tetepare training 78 Grassroots approach to Marine 30 Propagation techniques for Tubi Management 34 Phantoms of the forest 82 Conservation in Solomon Islands: acts without actions 38 Choiseul Island: Protecting Mt Cover page The newly discovered Vangunu Maetambe to Kolombangara River Island endemic rat, Uromys vika. Image watershed credit: Velizar Simeonovski, Field Museum. wildernesssolomons.com WWW.MELANESIANGEO.COM | 3 Melanesian EDITORS NOTE Geo PRODUCTION TEAM Government Of Founder/Editor: Patrick Pikacha of the priority species listed in the Critical Ecosystem [email protected] Solomon Islands Hails Partnership Fund’s investment strategy for the East Assistant editor: Tamara Osborne Melanesian Islands. [email protected] Barana Community The Critical Ecosystem Partnership Fund (CEPF) Contributing editor: David Boseto [email protected] is designed to safeguard Earth’s most biologically rich Prepress layout: Patrick Pikacha Nature Park Initiative and threatened regions, known as biodiversity hotspots.