Cryptosporidium Spp. in Wild and Captive Australian Flying Foxes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Novel Rhabdovirus Infecting Newly Discovered Nycteribiid Bat Flies
www.nature.com/scientificreports OPEN Kanyawara Virus: A Novel Rhabdovirus Infecting Newly Discovered Nycteribiid Bat Flies Received: 19 April 2017 Accepted: 25 May 2017 Infesting Previously Unknown Published: xx xx xxxx Pteropodid Bats in Uganda Tony L. Goldberg 1,2,3, Andrew J. Bennett1, Robert Kityo3, Jens H. Kuhn4 & Colin A. Chapman3,5 Bats are natural reservoir hosts of highly virulent pathogens such as Marburg virus, Nipah virus, and SARS coronavirus. However, little is known about the role of bat ectoparasites in transmitting and maintaining such viruses. The intricate relationship between bats and their ectoparasites suggests that ectoparasites might serve as viral vectors, but evidence to date is scant. Bat flies, in particular, are highly specialized obligate hematophagous ectoparasites that incidentally bite humans. Using next- generation sequencing, we discovered a novel ledantevirus (mononegaviral family Rhabdoviridae, genus Ledantevirus) in nycteribiid bat flies infesting pteropodid bats in western Uganda. Mitochondrial DNA analyses revealed that both the bat flies and their bat hosts belong to putative new species. The coding-complete genome of the new virus, named Kanyawara virus (KYAV), is only distantly related to that of its closest known relative, Mount Elgon bat virus, and was found at high titers in bat flies but not in blood or on mucosal surfaces of host bats. Viral genome analysis indicates unusually low CpG dinucleotide depletion in KYAV compared to other ledanteviruses and rhabdovirus groups, with KYAV displaying values similar to rhabdoviruses of arthropods. Our findings highlight the possibility of a yet- to-be-discovered diversity of potentially pathogenic viruses in bat ectoparasites. Bats (order Chiroptera) represent the second largest order of mammals after rodents (order Rodentia). -

Seasonal Shedding of Coronaviruses in Straw-Colored Fruit Bats at Urban Roosts in Africa
Seasonal Shedding of Coronaviruses in Straw-colored Fruit Bats at Urban Roosts in Africa The adaptation of bats (order Chiroptera) to use and For these reasons, we assessed the seasonality of occupy human dwellings across the planet has created coronavirus (CoV) shedding by the straw-colored intensive bat-human interfaces. Because bats provide fruit bat (Eidolon helvum) by passively collecting 97 important ecosystem services and also host and shed fecal samples on a monthly basis during a entire year zoonotic viruses, these interfaces represent a double in two urban colonies: Accra, Ghana (West Africa) challenge: i) the conservation of bats and their services and Morogoro, Tanzania (East Africa; Fig 1). Sampling and ii) the prevention of viral spillover. collection was conducted under the same trees during Many species of bats have evolved a seasonal life the study period. This species of fruit bat shows a single history that has resulted in the development of specific birth pulse during the year, its colonies show spectacular reproductive and foraging activities during distinctive periodical changes in size, and similarly to other tree- periods of the year. For example, many species mate, roosting megabats, several roosts are located in busy give birth, and nurse during particular and predictable urban centers across sub-Saharan Africa. Moreover, times of the year. Moreover, bat migration can produce we concomitantly collected data on the roost sizes predictable variations in colony sizes during a typical and precipitation levels over time, and we established year, from a complete absence of bats to the aggregation the reproductive periods through the year (birth of millions of individuals depending on the season. -

Zoologische Mededelingen
ZOOLOGISCHE MEDEDELINGEN UITGEGEVEN DOOR HET RIJKSMUSEUM VAN NATUURLIJKE HISTORIE TE LEIDEN (MINISTERIE VAN CULTUUR, RECREATIE EN MAATSCHAPPELIJK WERK) Deel 55 no. 14 4 maart 1980 A NEW FRUIT BAT OF THE GENUS MYONYCTERIS MATSCHIE, 1899, FROM EASTERN KENYA AND TANZANIA (MAMMALIA, MEGACHIROPTERA) by W. BERGMANS Instituut voor Taxonomische Zoölogie, Universiteit van Amsterdam With 4 text-figures ABSTRACT Myonycteris relicta n. sp. is described from the Shimba Hills in southeast Kenya and from the Usambara Mountains in northeast Tanzania. The species is larger than the only other known African mainland species of the genus, Myonycteris torquata (Dobson, 1878), from the Central and West African rain forests and, if compared to M. torquata and the only other species in the genus, M. brachycephala (Bocage, 1889) from São Tomé, has a relatively longer rostrum, a more deflected cranial axis, and further differs in number, shape and position of its teeth. The new species provides new arguments for the relationship between the genera Myonycteris Matschie, 1899, and Lissonycteris Andersen, 1912. It is believed that Myonycteris relicta may be a forest species and as such restricted to isolated East African forests. INTRODUCTION During a visit to the Zoologisches Museum in Berlin (ZMB), in April 1979, the author found two fruit bat specimens from the Tanzanian Usa- mbara Mountains, which proved to represent an undescribed taxon. Later, in June 1979, Dr C. Smeenk of the Rijksmuseum van Natuurlijke Historie at Leiden (RMNH) recognized a third specimen of this taxon in newly acquired material from the Shimba Hills in southeast Kenya. The bats differ on specific level from all other known fruit bats, and are described in the present paper. -
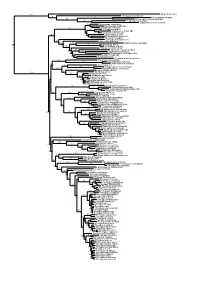
Figs1 ML Tree.Pdf
100 Megaderma lyra Rhinopoma hardwickei 71 100 Rhinolophus creaghi 100 Rhinolophus ferrumequinum 100 Hipposideros armiger Hipposideros commersoni 99 Megaerops ecaudatus 85 Megaerops niphanae 100 Megaerops kusnotoi 100 Cynopterus sphinx 98 Cynopterus horsfieldii 69 Cynopterus brachyotis 94 50 Ptenochirus minor 86 Ptenochirus wetmorei Ptenochirus jagori Dyacopterus spadiceus 99 Sphaerias blanfordi 99 97 Balionycteris maculata 100 Aethalops alecto 99 Aethalops aequalis Thoopterus nigrescens 97 Alionycteris paucidentata 33 99 Haplonycteris fischeri 29 Otopteropus cartilagonodus Latidens salimalii 43 88 Penthetor lucasi Chironax melanocephalus 90 Syconycteris australis 100 Macroglossus minimus 34 Macroglossus sobrinus 92 Boneia bidens 100 Harpyionycteris whiteheadi 69 Harpyionycteris celebensis Aproteles bulmerae 51 Dobsonia minor 100 100 80 Dobsonia inermis Dobsonia praedatrix 99 96 14 Dobsonia viridis Dobsonia peronii 47 Dobsonia pannietensis 56 Dobsonia moluccensis 29 Dobsonia anderseni 100 Scotonycteris zenkeri 100 Casinycteris ophiodon 87 Casinycteris campomaanensis Casinycteris argynnis 99 100 Eonycteris spelaea 100 Eonycteris major Eonycteris robusta 100 100 Rousettus amplexicaudatus 94 Rousettus spinalatus 99 Rousettus leschenaultii 100 Rousettus aegyptiacus 77 Rousettus madagascariensis 87 Rousettus obliviosus Stenonycteris lanosus 100 Megaloglossus woermanni 100 91 Megaloglossus azagnyi 22 Myonycteris angolensis 100 87 Myonycteris torquata 61 Myonycteris brachycephala 33 41 Myonycteris leptodon Myonycteris relicta 68 Plerotes anchietae -

Conventional Wisdom on Roosting Behaviour of Australian
Conventional wisdom on roosting behaviour of Australian flying foxes - a critical review, and evaluation using new data Tamika Lunn1, Peggy Eby2, Remy Brooks1, Hamish McCallum1, Raina Plowright3, Maureen Kessler4, and Alison Peel1 1Griffith University 2University of New South Wales 3Montana State University 4Montana State University System April 12, 2021 Abstract 1. Fruit bats (Family: Pteropodidae) are animals of great ecological and economic importance, yet their populations are threatened by ongoing habitat loss and human persecution. A lack of ecological knowledge for the vast majority of Pteropodid bat species presents additional challenges for their conservation and management. 2. In Australia, populations of flying-fox species (Genus: Pteropus) are declining and management approaches are highly contentious. Australian flying-fox roosts are exposed to management regimes involving habitat modification, either through human-wildlife conflict management policies, or vegetation restoration programs. Details on the fine-scale roosting ecology of flying-foxes are not sufficiently known to provide evidence-based guidance for these regimes and the impact on flying-foxes of these habitat modifications is poorly understood. 3. We seek to identify and test commonly held understandings about the roosting ecology of Australian flying-foxes to inform practical recommendations and guide and refine management practices at flying-fox roosts. 4. We identify 31 statements relevant to understanding of flying-fox roosting structure, and synthesise these in the context of existing literature. We then contribute contemporary data on the fine-scale roosting structure of flying-fox species in south-eastern Queensland and north- eastern New South Wales, presenting a 13-month dataset from 2,522 spatially referenced roost trees across eight sites. -

Final Report on the Project
BP Conservation Programme (CLP) 2005 PROJECT NO. 101405 - BRONZE AWARD WINNER ECOLOGY, DISTRIBUTION, STATUS AND PROTECTION OF THREE CONGOLESE FRUIT BATS FINAL REPORT Patrick KIPALU Team Leader Observatoire Congolais pour la Protection de l’Environnement OCPE – ong Kinshasa – Democratic Republic of the Congo E-mail: [email protected] APRIL 2009 1 Table of Content Acknowledgements…………………………………………………………………. p3 I. Project Summary……………………………………………………………….. p4 II. Introduction…………………………………………………………………… p4-p7 III. Materials and Methods ……………………………………………………….. p7-p10 IV. Results per Study Site…………………………………………………………. p10-p15 1. Pointe-Noire ………………………………………………………….. p10-p12 2. Mayumbe Forest /Luki Reserve……………………………………….. p12-p13 3. Zongo Forest…………………………………………………………... p14 4. Mbanza-Ngungu ………………………………………………………. P15 V. General Results ………………………………………………………………p15-p16 VI. Discussions……………………………………………………………………p17-18 VII. Conclusion and Recommendations……………………………………….p 18-p19 VIII. Bibliography………………………………………………………………p20-p21 Acknowledgements 2 The OCPE (Observatoire Congolais pour la Protection de l’Environnement) project team would like to start by expressing our gratefulness and saying thank you to the BP Conservation Program, which has funded the execution of this project. The OCPE also thanks the Van Tienhoven Foundation which provided a further financial support. Without these organisations, execution of the project would not have been possible. We would like to thank specially the BPCP “dream team”: Marianne D. Carter, Robyn Dalzen and our regretted Kate Stoke for their time, advices, expertise and care, which helped us to complete this work, Our special gratitude goes to Dr. Wim Bergmans, who was the hero behind the scene from the conception to the execution of the research work. Without his expertise, advices and network it would had been difficult for the project team to produce any result from this project. -

A Review of the Pteropus Rufus (É. Geoffroy, 1803) Colonies Within The
ARTICLE IN PRESS - EARLY VIEW MADAGASCAR CONSERVATION & DEVELOPMENT VOLUME 11 | ISSUE 1 — JUNE 2016 page 1 ARTICLE http://dx.doi.org/10.4314/mcd.v11i1.7 A review of the Pteropus rufus (É. Geoffroy, 1803) colonies within the Tolagnaro region of southeast Madagascar – an assessment of neoteric threats and conservation condition Sam Hyde RobertsI, Mark D. JacobsI, Ryan M. ClarkI, Correspondence: Charlotte M. DalyII, Longosoa H. TsimijalyIII, Retsiraiky J. Sam Hyde Roberts RossizelaIII, Samuel T. PrettymanI SEED Madagascar, Studio 7, 1A Beethoven Street, London W10 4LG, United Kingdom Email: [email protected] ABSTRACT soit parce qu’elles se sont déplacées suite à des dérangements. We surveyed 10 Pteropus rufus roost sites within the sou- Les effectifs d’une seule colonie semblent avoir diminué de theastern Anosy Region of Madagascar to provide an update on manière significative tandis que ceux de trois autres colonies the areas’ known flying fox population and its conservation status. semblent avoir été maintenus à leur niveau. Notre étude a montré We report on two new colonies from Manambaro and Mandena que l'abondance globale de P. rufus dans la région n’a augmenté and provide an account of the colonies first reported and last as- que d’un pourcent depuis 2006 et que cette augmentation était le sessed in 2006. Currently only a solitary roost site receives any résultat de la protection garantie au dortoir dans la réserve privée formal protection (Berenty) whereas further two colonies rely so- de Berenty. À la lumière d'un décret qui a imposé une période de lely on taboo ‘fady’ for their security. -

Index of Handbook of the Mammals of the World. Vol. 9. Bats
Index of Handbook of the Mammals of the World. Vol. 9. Bats A agnella, Kerivoula 901 Anchieta’s Bat 814 aquilus, Glischropus 763 Aba Leaf-nosed Bat 247 aladdin, Pipistrellus pipistrellus 771 Anchieta’s Broad-faced Fruit Bat 94 aquilus, Platyrrhinus 567 Aba Roundleaf Bat 247 alascensis, Myotis lucifugus 927 Anchieta’s Pipistrelle 814 Arabian Barbastelle 861 abae, Hipposideros 247 alaschanicus, Hypsugo 810 anchietae, Plerotes 94 Arabian Horseshoe Bat 296 abae, Rhinolophus fumigatus 290 Alashanian Pipistrelle 810 ancricola, Myotis 957 Arabian Mouse-tailed Bat 164, 170, 176 abbotti, Myotis hasseltii 970 alba, Ectophylla 466, 480, 569 Andaman Horseshoe Bat 314 Arabian Pipistrelle 810 abditum, Megaderma spasma 191 albatus, Myopterus daubentonii 663 Andaman Intermediate Horseshoe Arabian Trident Bat 229 Abo Bat 725, 832 Alberico’s Broad-nosed Bat 565 Bat 321 Arabian Trident Leaf-nosed Bat 229 Abo Butterfly Bat 725, 832 albericoi, Platyrrhinus 565 andamanensis, Rhinolophus 321 arabica, Asellia 229 abramus, Pipistrellus 777 albescens, Myotis 940 Andean Fruit Bat 547 arabicus, Hypsugo 810 abrasus, Cynomops 604, 640 albicollis, Megaerops 64 Andersen’s Bare-backed Fruit Bat 109 arabicus, Rousettus aegyptiacus 87 Abruzzi’s Wrinkle-lipped Bat 645 albipinnis, Taphozous longimanus 353 Andersen’s Flying Fox 158 arabium, Rhinopoma cystops 176 Abyssinian Horseshoe Bat 290 albiventer, Nyctimene 36, 118 Andersen’s Fruit-eating Bat 578 Arafura Large-footed Bat 969 Acerodon albiventris, Noctilio 405, 411 Andersen’s Leaf-nosed Bat 254 Arata Yellow-shouldered Bat 543 Sulawesi 134 albofuscus, Scotoecus 762 Andersen’s Little Fruit-eating Bat 578 Arata-Thomas Yellow-shouldered Talaud 134 alboguttata, Glauconycteris 833 Andersen’s Naked-backed Fruit Bat 109 Bat 543 Acerodon 134 albus, Diclidurus 339, 367 Andersen’s Roundleaf Bat 254 aratathomasi, Sturnira 543 Acerodon mackloti (see A. -

High Diversity of West African Bat Malaria Parasites and a Tight Link with Rodent Plasmodium Taxa
High diversity of West African bat malaria parasites and a tight link with rodent Plasmodium taxa Juliane Schaera,b,1,2, Susan L. Perkinsc,1,2, Jan Decherd, Fabian H. Leendertze, Jakob Fahrf,g, Natalie Weberh, and Kai Matuschewskia,i aParasitology Unit, Max Planck Institute for Infection Biology, 10117 Berlin, Germany; bMuseum für Naturkunde, Leibniz Institute for Research on Evolution and Biodiversity, 10115 Berlin, Germany; cSackler Institute for Comparative Genomics, American Museum of Natural History, New York, NY 10024; dSection of Mammals, Zoologisches Forschungsmuseum A. König, 53113 Bonn, Germany; eRobert Koch Institute, 13302 Berlin, Germany; fZoological Institute, Technische Universität Braunschweig, 38106 Braunschweig, Germany; gDepartment of Migration and Immuno-ecology, Max Planck Institute for Ornithology, 78315 Radolfzell, Germany; hInstitute of Experimental Ecology, University of Ulm, 89069 Ulm, Germany; and iInstitute of Biology, Humboldt University, 10117 Berlin, Germany Edited by Jitender P. Dubey, US Department of Agriculture, Beltsville, MD, and approved September 10, 2013 (received for review June 10, 2013) As the only volant mammals, bats are captivating for their high to the late 19th century (10). The corresponding vectors for most taxonomic diversity, for their vital roles in ecosystems—particularly as bat parasites remain unknown. Similarly, the phylogenetic rela- pollinators and insectivores—and, more recently, for their important tionships for the majority of these parasites remain enigmatic. roles in the maintenance and transmission of zoonotic viral diseases. Here, we present a unique systematic analysis of Haemosporida Genome sequences have identified evidence for a striking expansion in a diverse species assemblage of bats. For this study, we per- of and positive selection in gene families associated with immunity. -

Eidolon Helvum)
Niger. J. Physiol. Sci. 35 (December 2020): 181 - 186 www.njps.physiologicalsociety.com Research Article Haematological Studies and Micronucleus Assay of Straw-Coloured Fruit Bats (Eidolon helvum) Anosike F.1, Lanipekun D.O.1, Adebiyi O.F.2, Ogunsuyi O.M.3, Bakare A.A.3 and *Olopade J.O.1 1Department of Veterinary Anatomy, University of Ibadan, Ibadan, Nigeria. 2Department of Veterinary Biochemistry and Physiology, University of Ibadan, Ibadan, Nigeria. 3Cell Biology and Genetics Unit, Department of Zoology, University of Ibadan, Ibadan, Nigeria. Summary: The straw-coloured fruit bats (Eidolon helvum) are the most widely distributed megachiropteran species in Africa. Studies have shown that they migrate, and are likely exposed to environmental pollutants across population. This study was designed to investigate genotoxicity via the bone marrow micronucleus assay and haematological alterations of Eidolon helvum in the tropics. Healthy straw-coloured fruit bats (Eidolon helvum; n=20) were captured from two geographical regions, Ogun and Gombe States in Nigeria and were grouped based on sex and age. Blood samples were collected for haematology and osmotic fragility, and bone marrow samples for genotoxicity studies. Results showed no significant differences in erythrocytes and leucocytes values across age and sex. The erythrocytes osmotic fragility was higher in juvenile than in adults at 0 and 0.1%NaCl, while it was higher in adult males than in adult females at 0 and 0.3% NaCl. The erythrocytes and leucocytes parameters in straw-colored fruit bats were within the reference values seen in literature except the higher monocyte counts suggesting chronic inflammation. There were increased levels of micronucleated polychromatic erythrocytes and normochromatic erythrocytes in the straw-coloured fruit bats indicating genotoxicity and cytotoxicity, respectively. -

Friends of Bats
Friends of Bats newsletter Issue 115 December, 2014 Ku-ring-gai Flying-fox Reserve – Council adopts management plan options, December 2014 Nancy Pallin During 2014 the camp of grey-headed Prior to the meeting KBCS emailed all At the Council meeting two residents flying-foxes remained next to houses on Councillors with the following comments: spoke against the proposal, wanting it to the north side of Ku-ring-gai Flying-fox go further. KBCS Chair, Tim Pearson and Support for recommendations A, B and Reserve (KFFR). Residents close to the Nancy Pallin spoke for the proposal as a C, provided that each tree was assessed camp continued to find the noise, smell reasonable compromise, arguing against on a case-by-case basis. and droppings intolerable. They strongly any additional measures. Several other lobbied Councillors seeking a solution. Requests for a 100m buffer considered KBCS members attended. totally unacceptable as, for most of its Relocation of the release cage and tree Council resolved to endorse and/or fund length, the reserve is only 150m wide. removal/pruning previously undertaken the management actions A, B and C were unsuccessful in reducing the Support financial assistance to above, aimed at “nudging” flying-foxes impacts on residents. double-glaze and insulate homes to from properties adjacent to KFFR. The financial costs of the various options In October, the Mayor, a staff member, reduce the impact of noise. weighed heavily on the Councillors. two residents and the Member for The risk of contracting diseases from There was considerable debate amongst Davidson met the NSW Minister for flying-foxes is extremely low. -

Interrogating Phylogenetic Discordance Resolves Deep Splits
Interrogating Phylogenetic Discordance Resolves Deep Splits in the Rapid Radiation of Old World Fruit Bats (Chiroptera: Pteropodidae) Nicolas Nesi, Georgia Tsagkogeorga, Susan Tsang, Violaine Nicolas, Aude Lalis, Annette Scanlon, Silke Riesle-Sbarbaro, Sigit Wiantoro, Alan Hitch, Javier Juste, et al. To cite this version: Nicolas Nesi, Georgia Tsagkogeorga, Susan Tsang, Violaine Nicolas, Aude Lalis, et al.. Interrogat- ing Phylogenetic Discordance Resolves Deep Splits in the Rapid Radiation of Old World Fruit Bats (Chiroptera: Pteropodidae). Systematic Biology, Oxford University Press (OUP), 2021, 10.1093/sys- bio/syab013. hal-03167896 HAL Id: hal-03167896 https://hal.sorbonne-universite.fr/hal-03167896 Submitted on 12 Mar 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Interrogating Phylogenetic Discordance Resolves Deep Splits in the Rapid Radiation of Old World Fruit Bats (Chiroptera: Pteropodidae) Downloaded from https://academic.oup.com/sysbio/advance-article/doi/10.1093/sysbio/syab013/6161783 by Sorbonne Université user on 12 March 2021 NICOLAS NESI1, GEORGIA TSAGKOGEORGA1, SUSAN M. TSANG2,3, VIOLAINE NICOLAS4, AUDE LALIS4, ANNETTE T. SCANLON5, SILKE A. RIESLE- SBARBARO6,7†, SIGIT WIANTORO8, ALAN T. HITCH9, JAVIER JUSTE10, CORINNA A. PINZARI11, FRANK J. BONACCORSO12, CHRISTOPHER M.