Epigenetic Regulations of Ahr in the Aspect of Immunomodulation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Aryl Hydrocarbon Receptor: Structural Analysis and Activation Mechanisms
The Aryl Hydrocarbon Receptor: Structural Analysis and Activation Mechanisms This thesis is submitted in fulfilment of the requirements for the degree of Doctor of Philosophy in the School of Molecular and Biomedical Sciences (Biochemistry), The University of Adelaide, Australia Fiona Whelan, B.Sc. (Hons) 2009 2 Table of Contents THESIS SUMMARY................................................................................. 6 DECLARATION....................................................................................... 7 PUBLICATIONS ARISING FROM THIS THESIS.................................... 8 ACKNOWLEDGEMENTS...................................................................... 10 ABBREVIATIONS ................................................................................. 12 CHAPTER 1: INTRODUCTION ............................................................. 17 1.1 BHLH.PAS PROTEINS ............................................................................................17 1.1.1 General background..................................................................................17 1.1.2 bHLH.PAS Class I Proteins.........................................................................18 1.2 THE ARYL HYDROCARBON RECEPTOR......................................................................19 1.2.1 Domain Structure and Ligand Activation ..............................................19 1.2.2 AhR Expression and Developmental Activity .......................................21 1.2.3 Mouse AhR Knockout Phenotype ...........................................................23 -

Open Thesis Master Document V5.0.Pdf
The Pennsylvania State University The Graduate School Department of Veterinary and Biomedical Science IDENTIFICATION OF ENDOGENOUS MODULATORS FOR THE ARYL HYDROCARBON RECEPTOR A Thesis in Genetics by Christopher R. Chiaro © 2007 Christopher R. Chiaro Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy December, 2007 The thesis of Christopher R. Chiaro was reviewed and approved* by the following: Gary H. Perdew John T. and Paige S. Smith Professor in Agricultural Sciences Thesis Advisor Chair of Committee C. Channa Reddy Distinguished Professor of Veterinary Science A. Daniel Jones Senior Scientist Department of Chemistry John P. Vanden Heuvel Professor of Veterinary Science Richard Ordway Associate Professor of Biology Chair of Genetics Graduate Program *Signatures are on file in the Graduate School iii ABSTRACT The aryl hydrocarbon receptor (AhR) is a ligand-activated transcription factor capable of being regulated by a structurally diverse array of chemicals ranging from environmental carcinogens to dietary metabolites. A member of the basic helix-loop- helix/ Per-Arnt-Sim (bHLH-PAS) super-family of DNA binding regulatory proteins, the AhR is an important developmental regulator that can be detected in nearly all mammalian tissues. Prior to ligand activation, the AhR resides in the cytosol as part of an inactive oligomeric protein complex comprised of the AhR ligand-binding subunit, a dimer of the 90 kDa heat shock protein, and a single molecule each of the immunophilin like X-associated protein 2 (XAP2) and p23 proteins. Functioning as chemosensor, the AhR responds to both endobiotic and xenobiotic derived chemical ligands by ultimately directing the expression of metabolically important target genes. -

Environmental Pollution 275 (2021) 116665
Environmental Pollution 275 (2021) 116665 Contents lists available at ScienceDirect Environmental Pollution journal homepage: www.elsevier.com/locate/envpol Metabolomics reveals the reproductive abnormality in female zebrafish exposed to environmentally relevant levels of climbazole* Ting Zou a, 1, Yan-Qiu Liang b, 1, Xiaoliang Liao a, Xiao-Fan Chen a, Tao Wang c, * Yuanyuan Song c, Zhi-Cheng Lin a, Zenghua Qi a, Zhi-Feng Chen a, d, , Zongwei Cai a, c a Guangdong Key Laboratory of Environmental Catalysis and Health Risk Control, School of Environmental Science and Engineering, Institute of Environmental Health and Pollution Control, Guangdong University of Technology, Guangzhou, 510006, China b Faculty of Chemistry and Environmental Science, Guangdong Ocean University, Zhanjiang, 524088, China c State Key Laboratory of Environmental and Biological Analysis, Department of Chemistry, Hong Kong Baptist University, Hong Kong Special Administrative Region, China d Guangdong Provincial Key Laboratory of Chemical Pollution and Environmental Safety, South China Normal University, Guangzhou, 510006, China article info abstract Article history: Climbazole (CBZ) ubiquitously detected in the aquatic environment may disrupt fish reproductive Received 15 November 2020 function. Thus far, the previous study has focused on its transcriptional impact of steroidogenesis-related Received in revised form genes on zebrafish, but the underlying toxic mechanism still needs further investigation at the metabolic 10 January 2021 level. In this study, adult zebrafish were chronically exposed to CBZ at concentrations of 0.1 (corre- Accepted 2 February 2021 sponding to the real concentration in surface water), 10, and 1000 mg/L and evaluated for reproductive Available online 5 February 2021 function by egg production, with subsequent ovarian tissue samples taken for histology, metabolomics, and other biochemical analysis. -

Role of Arachidonic Acid and Its Metabolites in the Biological and Clinical Manifestations of Idiopathic Nephrotic Syndrome
International Journal of Molecular Sciences Review Role of Arachidonic Acid and Its Metabolites in the Biological and Clinical Manifestations of Idiopathic Nephrotic Syndrome Stefano Turolo 1,* , Alberto Edefonti 1 , Alessandra Mazzocchi 2, Marie Louise Syren 2, William Morello 1, Carlo Agostoni 2,3 and Giovanni Montini 1,2 1 Fondazione IRCCS Ca’ Granda-Ospedale Maggiore Policlinico, Pediatric Nephrology, Dialysis and Transplant Unit, Via della Commenda 9, 20122 Milan, Italy; [email protected] (A.E.); [email protected] (W.M.); [email protected] (G.M.) 2 Department of Clinical Sciences and Community Health, University of Milan, 20122 Milan, Italy; [email protected] (A.M.); [email protected] (M.L.S.); [email protected] (C.A.) 3 Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Pediatric Intermediate Care Unit, 20122 Milan, Italy * Correspondence: [email protected] Abstract: Studies concerning the role of arachidonic acid (AA) and its metabolites in kidney disease are scarce, and this applies in particular to idiopathic nephrotic syndrome (INS). INS is one of the most frequent glomerular diseases in childhood; it is characterized by T-lymphocyte dysfunction, alterations of pro- and anti-coagulant factor levels, and increased platelet count and aggregation, leading to thrombophilia. AA and its metabolites are involved in several biological processes. Herein, Citation: Turolo, S.; Edefonti, A.; we describe the main fields where they may play a significant role, particularly as it pertains to their Mazzocchi, A.; Syren, M.L.; effects on the kidney and the mechanisms underlying INS. AA and its metabolites influence cell Morello, W.; Agostoni, C.; Montini, G. -

Lipoxin A4: a Novel Anti-Inflammatory Molecule? Thorax: First Published As 10.1136/Thx.50.2.111 on 1 February 1995
Thorax 1995;50:111-112 illl Lipoxin A4: a novel anti-inflammatory molecule? Thorax: first published as 10.1136/thx.50.2.111 on 1 February 1995. Downloaded from Arachidonic acid is metabolised by the cyclooxygenase contractions.22 These studies support the view that LXA4 pathway to the prostaglandins and thromboxane A2 or via may act as a partial agonist at the same or similar site as one of the lipoxygenase pathways.' Three major lip- the sulphidopeptide leukotrienes. oxygenase pathways have been identified in mammalian The fact that 1 5-lipoxygenase is abundant in lung tissue tissue - namely, the 5-, 12-, and 15-lipoxygenases.2A The and that LXA4 has been recovered in the bronchoalveolar 5-lipoxygenase pathway metabolises arachidonic acid lavage fluid ofpatients with asthma and other lung diseases through two intermediates, 5-hydroperoxyeicosatetranoic suggests that LXA4 may be a potential mediator or mod- acid (5-HPETE) and leukotriene A4 (LTA4), to LTB4 and ulator of inflammation in the lung. In a recent study eight the sulphidopeptide leukotrienes LTC4, LTD4, and LTE4.5 subjects underwent inhalation challenge with LXA4,23 but The sulphidopeptide leukotrienes are potent spasmogens6 no effect was seen on specific conductance, rate of airflow for non-vascular smooth muscle and may play a part in at 25% vital capacity (V25), blood pressure, pulse, or the pathogenesis of bronchial asthma.7`10 asthmatic symptoms. There was, however, a significant The interactions between 5-lipoxygenase and 15-lip- shift of the specific conductance and V25 dose-response oxygenase on arachidonic acid metabolism have recently curve to the right after inhalation challenge with LTC4 been studied and a new series of biologically active me- combined with LXA4 compared with that after inhalation tabolites described." 12 These molecules have been termed challenge with LTC4 alone. -
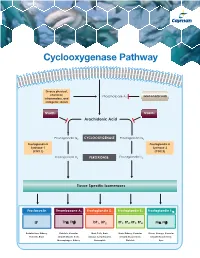
Cyclooxygenase Pathway
Cyclooxygenase Pathway Diverse physical, chemical, Phospholipase A Glucocorticoids inflammatory, and 2 mitogenic stimuli NSAIDs NSAIDs Arachidonic Acid Prostaglandin G2 CYCLOOXYGENASE Prostaglandin G2 Prostaglandin H Prostaglandin H Synthase-1 Synthase-2 (COX 1) (COX 2) Prostaglandin H2 PEROXIDASE Prostaglandin H2 Tissue Specific Isomerases Prostacyclin Thromboxane A2 Prostaglandin D2 Prostaglandin E2 Prostaglandin F2α IP TPα, TPβ DP1, DP2 EP1, EP2, EP3, EP4 FPα, FPβ Endothelium, Kidney, Platelets, Vascular Mast Cells, Brain, Brain, Kidney, Vascular Uterus, Airways, Vascular Platelets, Brain Smooth Muscle Cells, Airways, Lymphocytes, Smooth Muscle Cells, Smooth Muscle Cells, Macrophages, Kidney Eosinophils Platelets Eyes Prostacyclin Item No. Product Features Prostacyclin (Prostaglandin I2; PGI2) is formed from arachidonic acid primarily in the vascular endothelium and renal cortex by sequential 515211 6-keto • Sample Types: Culture Medium | Plasma Prostaglandin • Measure 6-keto PGF levels down to 6 pg/ml activities of COX and prostacyclin synthase. PGI2 is non-enzymatically 1α F ELISA Kit • Incubation : 18 hours | Development: 90-120 minutes | hydrated to 6-keto PGF1α (t½ = 2-3 minutes), and then quickly converted 1α Read: Colorimetric at 405-420 nm to the major metabolite, 2,3-dinor-6-keto PGF1α (t½= 30 minutes). Prostacyclin was once thought to be a circulating hormone that regulated • Assay 24 samples in triplicate or 36 samples in duplicate platelet-vasculature interactions, but the rate of secretion into circulation • NOTE: A portion of urinary 6-keto PGF1α is of renal origin coupled with the short half-life indicate that prostacyclin functions • NOTE : It has been found that normal plasma levels of 6-keto PGF may be low locally. -

Reduced 15-Lipoxygenase 2 and Lipoxin A4/Leukotriene B4 Ratio in Children with Cystic Fibrosis
ORIGINAL ARTICLE CYSTIC FIBROSIS Reduced 15-lipoxygenase 2 and lipoxin A4/leukotriene B4 ratio in children with cystic fibrosis Fiona C. Ringholz1, Paul J. Buchanan1, Donna T. Clarke1, Roisin G. Millar1, Michael McDermott2, Barry Linnane1,3,4, Brian J. Harvey5, Paul McNally1,2 and Valerie Urbach1,6 Affiliations: 1National Children’s Research Centre, Crumlin, Dublin, Ireland. 2Our Lady’s Children’s Hospital, Crumlin, Dublin, Ireland. 3Midwestern Regional Hospital, Limerick, Ireland. 4Centre for Interventions in Infection, Inflammation and Immunity (4i), Graduate Entry Medical School, University of Limerick, Limerick, Ireland. 5Molecular Medicine Laboratories, Royal College of Surgeons in Ireland, Beaumont Hospital, Dublin, Ireland. 6Institut National de la Sante´ et de la Recherche Me´dicale, U845, Faculte´ de Me´decine Paris Descartes, Paris, France. Correspondence: Valerie Urbach, National Children’s Research Centre, Crumlin, Dublin 12, Ireland. E-mail: [email protected] ABSTRACT Airway disease in cystic fibrosis (CF) is characterised by impaired mucociliary clearance, persistent bacterial infection and neutrophilic inflammation. Lipoxin A4 (LXA4) initiates the active resolution of inflammation and promotes airway surface hydration in CF models. 15-Lipoxygenase (LO) plays a central role in the ‘‘class switch’’ of eicosanoid mediator biosynthesis from leukotrienes to lipoxins, initiating the active resolution of inflammation. We hypothesised that defective eicosanoid mediator class switching contributes to the failure to resolve inflammation in CF lung disease. Using bronchoalveolar lavage (BAL) samples from 46 children with CF and 19 paediatric controls we demonstrate that the ratio of LXA4 to leukotriene B4 (LTB4) is depressed in CF BAL (p,0.01), even in the absence of infection (p,0.001). -

Strict Regio-Specificity of Human Epithelial 15-Lipoxygenase-2
Strict Regio-specificity of Human Epithelial 15-Lipoxygenase-2 Delineates its Transcellular Synthesis Potential Abigail R. Green, Shannon Barbour, Thomas Horn, Jose Carlos, Jevgenij A. Raskatov, Theodore R. Holman* Department Chemistry and Biochemistry, University of California Santa Cruz, 1156 High Street, Santa Cruz CA 95064, USA *Corresponding author: Tel: 831-459-5884. Email: [email protected] FUNDING: This work was supported by the NIH NS081180 and GM56062. Abbreviations: LOX, lipoxygenase; h15-LOX-2, human epithelial 15-lipoxygenase-2; h15-LOX-1, human reticulocyte 15-lipoxygenase-1; sLO-1, soybean lipoxygenase-1; 5-LOX, leukocyte 5-lipoxygenase; 12-LOX, human platelet 12-lipoxygenase; GP, glutathione peroxidase; AA, arachidonic acid; HETE, hydoxy-eicosatetraenoic acid; HPETE, hydroperoxy-eicosatetraenoic acid; diHETEs, dihydroxy-eicosatetraenoic acids; 5-HETE, 5-hydroxy-6E,8Z,11Z,14Z-eicosatetraenoic acid; 5-HPETE, 5-hydro peroxy-6E,8Z,11Z,14Z-eicosatetraenoic acid; 12-HPETE, 12-hydroperoxy-5Z,8Z,10E, 14Z-eicosatetraenoic acid; 15-HPETE, 15-hydroperoxy-5Z,8Z,10Z,13E- eicosatetraenoic acid; 5,15-HETE, 5S,15S-dihydroxy-6E,8Z,10Z,13E-eicosatetraenoic acid; 5,15-diHPETE, 5,15-dihydroperoxy-6E,8Z,10Z,13E-eicosatetraenoic acid; 5,6- diHETE, 5S,6R-dihydroxy-7E,9E,11Z,14Z-eicosatetraenoic acid; LTA4, 5S-trans-5,6- oxido-7E,9E,11Z,14Z-eicosatetraenoic acid; LTB4, 5S,12R-dihydroxy-6Z,8E,10E,14Z- eicosatetraenoic acid; LipoxinA4 (LxA4), 5S,6R,15S-trihydroxy-7E,9E,11Z,13E- eicosatetraenoic acid; LipoxinB4 (LxB4), 5S,14R,15S-trihydroxy-6E,8Z,10E,12E- eicosatetraenoic acid. Abstract Lipoxins are an important class of lipid mediators that induce the resolution of inflammation, and arise from transcellular exchange of arachidonic acid (AA)- derived lipoxygenase products. -

Phospholipase A2 Regulates Eicosanoid Class Switching During Inflammasome Activation
Phospholipase A2 regulates eicosanoid class switching during inflammasome activation Paul C. Norrisa, David Gosselinb, Donna Reichartb, Christopher K. Glassb, and Edward A. Dennisa,1 Departments of aChemistry/Biochemistry and Pharmacology, and bCellular and Molecular Medicine, University of California, San Diego, La Jolla, CA 92093 Edited by Michael A. Marletta, The Scripps Research Institute, La Jolla, CA, and approved July 30, 2014 (received for review March 13, 2014) Initiation and resolution of inflammation are considered to be to initiate pathogenic killing, subsequent “class switching” to lipoxin tightly connected processes. Lipoxins (LX) are proresolution lipid (LX) formation by “reprogrammed” neutrophils inhibits additional mediators that inhibit phlogistic neutrophil recruitment and pro- neutrophil recruitment during self-resolving inflammatory resolu- mote wound-healing macrophage recruitment in humans via tion (9). The direct link between inflammatory commitment and potent and specific signaling through the LXA4 receptor (ALX). resolution mediated by eicosanoid signaling in macrophages One model of lipoxin biosynthesis involves sequential metabolism remains unclear from short-term vs. long-term priming, but the of arachidonic acid by two cell types expressing a combined trans- complete temporal changes and important interconnections cellular metabolon. It is currently unclear how lipoxins are effi- within the entire eicosadome are now demonstrated. ciently formed from precursors or if they are directly generated after receptor-mediated inflammatory commitment. Here, we pro- Results vide evidence for a pathway by which lipoxins are generated in We first primed immortalized macrophage-like cells (RAW264.7) macrophages as a consequence of sequential activation of toll-like with the TLR4 agonist Kdo2 lipid A (KLA) for various times and receptor 4 (TLR4), a receptor for endotoxin, and P2X7, a purinergic examined the effects on subsequent purinergic stimulated COX receptor for extracellular ATP. -

Identification of Potential Aryl Hydrocarbon Receptor Ligands by Virtual Screening of Industrial Chemicals
Chemical Research in Toxicology This document is confidential and is proprietary to the American Chemical Society and its authors. Do not copy or disclose without written permission. If you have received this item in error, notify the sender and delete all copies. Identification of potential aryl hydrocarbon receptor ligands by virtual screening of industrial chemicals Journal: Chemical Research in Toxicology Manuscript ID tx-2016-004609 Manuscript Type: Article Date Submitted by the Author: 20-Dec-2016 Complete List of Authors: Larsson, Malin; Umeå University, Department of Chemistry Fraccalvieri, Domenico; Università degli Studi di Milano-Bicocca, 1Dipartimento di Scienze dell’Ambiente e del Territorio Andersson, C. David; Umeå University, Department of Chemistry Bonati, Laura; University of Milano-Bicocca, Department of Earth and Environmental Sciences Linusson, Anna; Umeå University, Department of Chemistry Andersson, Patrik; Umea University, Department of Chemistry ACS Paragon Plus Environment Page 1 of 52 Chemical Research in Toxicology 1 2 3 4 Identification of potential aryl hydrocarbon receptor ligands by virtual 5 6 screening of industrial chemicals 7 8 9 10 Malin Larsson†, Domenico Fraccalvieri‡, C. David Andersson†, Laura Bonati‡, Anna Linusson†, and 11 12 Patrik L. Andersson†* 13 14 15 †Department of Chemistry, Umeå University, SE-901 87 Umeå, Sweden 16 17 18 ‡Department of Earth and Environmental Sciences, University of Milano-Bicocca, Piazza della Scienza 1, 19 20 21 20126 Milano, Italy 22 23 24 *Corresponding author: Tel.: +46-90-786-5266; fax: +46-90-786-7655. Email address: [email protected] 25 26 27 28 29 30 Malin Larsson†: [email protected] 31 32 33 ‡ 34 Domenico Fraccalvieri : [email protected] 35 36 † 37 C. -

The Protective Effects of Conjugated Linoleic Acid Against Carcinogenesis
The Protective Effects of Conjugated Linoleic Acid Against Carcinogenesis Item Type text; Electronic Dissertation Authors Kemp, Michael Quentin Publisher The University of Arizona. Rights Copyright © is held by the author. Digital access to this material is made possible by the University Libraries, University of Arizona. Further transmission, reproduction or presentation (such as public display or performance) of protected items is prohibited except with permission of the author. Download date 24/09/2021 15:13:15 Link to Item http://hdl.handle.net/10150/193637 THE PROTECTIVE EFFECTS OF CONJUGATED LINOLEIC ACID AGAINST CARCINOGENESIS by Michael Quentin Kemp A Dissertation Submitted to the Faculty of the GRADUATE PROGRAM IN NUTRITIONAL SCIENCES In Partial Fulfillment of the Requirements for the Degree of DOC TORAL OF PHILOSOPHY In the Graduate College THE UNIVERSITY OF ARIZONA 2005 2 THE UNIVERSITY OF ARIZONA GRADUATE COLLEGE As members of the Dissertation Committee, we certify that we have read the dissertation prepared by Michael Q. Kemp entitled The Protective Effec ts of Conjugated Linoleic Acid Against Carcinogenesis and recommend that it be accepted as fulfilling the dissertation requirement for the Deg ree of Doctor of Philosophy _______ ___________________________ __________________ ________ _______ ___ _ Date : 11/18/05 . Donato Romagnolo _______ ___________________________ __________________ ________ ___________ Date: 11/18/05 . Wanda Howell _______ ___________________________ __________________ ________ ___________ Date: 11/18/05 . Linda Houtkooper _______ ___________________________ __________________ ________ ___________ Date: 11/18/05 . Scott Going _______ ___________________________ __________________ ________ ___________ Date: 11/18/05 . Joy Winzerling Final approval and acceptance of this dissertation is contingent upon the candidate’s submission of the final copies of the dissertation to the Graduate College. -

Therapeutic Effects of Specialized Pro-Resolving Lipids Mediators On
antioxidants Review Therapeutic Effects of Specialized Pro-Resolving Lipids Mediators on Cardiac Fibrosis via NRF2 Activation 1, 1,2, 2, Gyeoung Jin Kang y, Eun Ji Kim y and Chang Hoon Lee * 1 Lillehei Heart Institute, University of Minnesota, Minneapolis, MN 55455, USA; [email protected] (G.J.K.); [email protected] (E.J.K.) 2 College of Pharmacy, Dongguk University, Seoul 04620, Korea * Correspondence: [email protected]; Tel.: +82-31-961-5213 Equally contributed. y Received: 11 November 2020; Accepted: 9 December 2020; Published: 10 December 2020 Abstract: Heart disease is the number one mortality disease in the world. In particular, cardiac fibrosis is considered as a major factor causing myocardial infarction and heart failure. In particular, oxidative stress is a major cause of heart fibrosis. In order to control such oxidative stress, the importance of nuclear factor erythropoietin 2 related factor 2 (NRF2) has recently been highlighted. In this review, we will discuss the activation of NRF2 by docosahexanoic acid (DHA), eicosapentaenoic acid (EPA), and the specialized pro-resolving lipid mediators (SPMs) derived from polyunsaturated lipids, including DHA and EPA. Additionally, we will discuss their effects on cardiac fibrosis via NRF2 activation. Keywords: cardiac fibrosis; NRF2; lipoxins; resolvins; maresins; neuroprotectins 1. Introduction Cardiovascular disease is the leading cause of death worldwide [1]. Cardiac fibrosis is a major factor leading to the progression of myocardial infarction and heart failure [2]. Cardiac fibrosis is characterized by the net accumulation of extracellular matrix proteins in the cardiac stroma and ultimately impairs cardiac function [3]. Therefore, interest in substances with cardioprotective activity continues.