Identifying Robust Biomarkers of Infection Through an Omics-Based
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
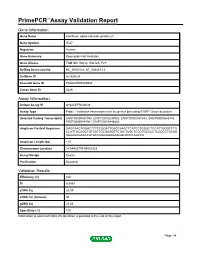
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name interferon, alpha-inducible protein 27 Gene Symbol IFI27 Organism Human Gene Summary Description Not Available Gene Aliases FAM14D, ISG12, ISG12A, P27 RefSeq Accession No. NC_000014.8, NT_026437.12 UniGene ID Hs.532634 Ensembl Gene ID ENSG00000165949 Entrez Gene ID 3429 Assay Information Unique Assay ID qHsaCEP0024638 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000557098, ENST00000298902, ENST00000557634, ENST00000555744, ENST00000444961, ENST00000448882 Amplicon Context Sequence GAGCAACTGGACTCTCCGGATTGACCAAGTTCATCCTGGGCTCCATTGGGTCTG CCATTGCGGCTGTCATTGCGAGGTTCTACTAGCTCCCTGCCCCTCGCCCTGCAG AGAAGAGAACCATGCCAGGGGAGAAGGCACCCAGCCA Amplicon Length (bp) 115 Chromosome Location 14:94582779-94582923 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 100 R2 0.9997 cDNA Cq 23.09 cDNA Tm (Celsius) 86 gDNA Cq 23.06 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report IFI27, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Human Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

IFI27 May Predict and Evaluate the Severity of Respiratory Syncytial Virus
Gao et al. Hereditas (2021) 158:3 https://doi.org/10.1186/s41065-020-00167-5 RESEARCH Open Access IFI27 may predict and evaluate the severity of respiratory syncytial virus infection in preterm infants Junyan Gao1, Xueping Zhu2, Mingfu Wu1, Lijun Jiang1, Fudong Wang1 and Shan He2,3* Abstract Background: Preterm infants are a special population that vulnerable to respiratory syncytial virus (RSV) infection and the lower respiratory tract infections (LRTIs) caused by RSV could be severe and even life-threating. The purpose of the present study was to identify candidate genes of preterm infants who are susceptible to RSV infection and provide a new insight into the pathogenesis of RSV infection. Methods: Three datasets (GSE77087, GSE69606 and GSE41374) containing 183 blood samples of RSV infected patients and 33 blood samples of healthy controls from Gene Expression Omnibus (GEO) database were downloaded and the differentially expressed genes (DEGs) were screened out. The function and pathway enrichments were analyzed through Database for Annotation, Visualization and Integrated Discovery (DAVID) website. The protein-protein interaction (PPI) network for DEGs was constructed through Search Tool for the Retrieval of Interacting Genes (STRING). The module analysis was performed by Cytoscape software and hub genes were identified. Clinical verification was employed to verify the expression level of top five hub genes among 72 infants including 50 RSV infected patients and 22 non-RSV-infected patients hospitalized in our center. Further, the RSV infected infants with high-expression IFI27 and those with low-expression IFI27 were compared (defined as higher or lower than the median mRNA level). -

IFI27 May Predict and Evaluate the Severity of Respiratory Syncytial Virus Infection in Preterm Infants
IFI27 May Predict and Evaluate the Severity of Respiratory Syncytial Virus Infection in Preterm Infants Junyan Gao Aliated Hospital of Yangzhou University https://orcid.org/0000-0001-6678-6147 Xueping Zhu Children's Hospital of Soochow University Mingfu Wu Aliated Hospital of Yangzhou University Lijun Jiang Aliated Hospital of Yangzhou University Fudong Wang Aliated Hospital of Yangzhou University Shan He ( [email protected] ) Research Keywords: respiratory syncytial virus, preterm infants, differentially expressed genes, protein‐protein interaction, pathway enrichment, interferon alpha inducible protein 27 Posted Date: July 15th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-42880/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published on January 2nd, 2021. See the published version at https://doi.org/10.1186/s41065-020-00167-5. Page 1/21 Abstract Background: Preterm infants are a special population that vulnerable to respiratory syncytial virus (RSV) infection and the lower respiratory tract infection (LRTIs) caused by RSV could be severe and even life- threating. The purpose of the present study was to identify candidate genes of preterm infants who are susceptible to RSV infection and provide a new insight into the pathogenesis of RSV infection. Methods: Three datasets (GSE77087, GSE69606 and GSE41374) containing 183 blood samples of RSV infected patients and 33 blood samples of healthy controls from Gene Expression Omnibus (GEO) database were downloaded and the differentially expressed genes (DEGs) were screened out. The function and pathway enrichments were analyzed through Database for Annotation, Visualization and Integrated Discovery (DAVID) website. -

Identification of Key Genes and Pathways in Pancreatic Cancer
G C A T T A C G G C A T genes Article Identification of Key Genes and Pathways in Pancreatic Cancer Gene Expression Profile by Integrative Analysis Wenzong Lu * , Ning Li and Fuyuan Liao Department of Biomedical Engineering, College of Electronic and Information Engineering, Xi’an Technological University, Xi’an 710021, China * Correspondence: [email protected]; Tel.: +86-29-86173358 Received: 6 July 2019; Accepted: 7 August 2019; Published: 13 August 2019 Abstract: Background: Pancreatic cancer is one of the malignant tumors that threaten human health. Methods: The gene expression profiles of GSE15471, GSE19650, GSE32676 and GSE71989 were downloaded from the gene expression omnibus database including pancreatic cancer and normal samples. The differentially expressed genes between the two types of samples were identified with the Limma package using R language. The gene ontology functional and pathway enrichment analyses of differentially-expressed genes were performed by the DAVID software followed by the construction of a protein–protein interaction network. Hub gene identification was performed by the plug-in cytoHubba in cytoscape software, and the reliability and survival analysis of hub genes was carried out in The Cancer Genome Atlas gene expression data. Results: The 138 differentially expressed genes were significantly enriched in biological processes including cell migration, cell adhesion and several pathways, mainly associated with extracellular matrix-receptor interaction and focal adhesion pathway in pancreatic cancer. The top hub genes, namely thrombospondin 1, DNA topoisomerase II alpha, syndecan 1, maternal embryonic leucine zipper kinase and proto-oncogene receptor tyrosine kinase Met were identified from the protein–protein interaction network. -

Mouse Ifi27l2a Conditional Knockout Project (CRISPR/Cas9)
https://www.alphaknockout.com Mouse Ifi27l2a Conditional Knockout Project (CRISPR/Cas9) Objective: To create a Ifi27l2a conditional knockout Mouse model (C57BL/6J) by CRISPR/Cas-mediated genome engineering. Strategy summary: The Ifi27l2a gene (NCBI Reference Sequence: NM_029803 ; Ensembl: ENSMUSG00000079017 ) is located on Mouse chromosome 12. 4 exons are identified, with the ATG start codon in exon 1 and the TGA stop codon in exon 4 (Transcript: ENSMUST00000055071). Exon 3 will be selected as conditional knockout region (cKO region). Deletion of this region should result in the loss of function of the Mouse Ifi27l2a gene. To engineer the targeting vector, homologous arms and cKO region will be generated by PCR using BAC clone RP24-62E20 as template. Cas9, gRNA and targeting vector will be co-injected into fertilized eggs for cKO Mouse production. The pups will be genotyped by PCR followed by sequencing analysis. Note: Mice homozygous for a knock-out allele exhibit normal susceptibility to infection with H1N1 or H7N7 influenza A virus, although sloughing of dead cells into bronchiolar lumen persists somewhat longer after infection than in infected controls. Exon 3 is not frameshift exon, and covers 60.0% of the coding region. The size of intron 2 for 5'-loxP site insertion: 488 bp, and the size of intron 3 for 3'-loxP site insertion: 448 bp. The size of effective cKO region: ~630 bp. The cKO region does not have any other known gene. Page 1 of 7 https://www.alphaknockout.com Overview of the Targeting Strategy Wildtype allele gRNA region 5' gRNA region 3' 1 2 3 4 Targeting vector Targeted allele Constitutive KO allele (After Cre recombination) Legends Homology arm Exon of mouse Ifi27l2a cKO region loxP site Page 2 of 7 https://www.alphaknockout.com Overview of the Dot Plot Window size: 10 bp Forward Reverse Complement Sequence 12 Note: The sequence of homologous arms and cKO region is aligned with itself to determine if there are tandem repeats. -

Supplementary Data
Supplemental figures Supplemental figure 1: Tumor sample selection. A total of 98 thymic tumor specimens were stored in Memorial Sloan-Kettering Cancer Center tumor banks during the study period. 64 cases corresponded to previously untreated tumors, which were resected upfront after diagnosis. Adjuvant treatment was delivered in 7 patients (radiotherapy in 4 cases, cyclophosphamide- doxorubicin-vincristine (CAV) chemotherapy in 3 cases). 34 tumors were resected after induction treatment, consisting of chemotherapy in 16 patients (cyclophosphamide-doxorubicin- cisplatin (CAP) in 11 cases, cisplatin-etoposide (PE) in 3 cases, cisplatin-etoposide-ifosfamide (VIP) in 1 case, and cisplatin-docetaxel in 1 case), in radiotherapy (45 Gy) in 1 patient, and in sequential chemoradiation (CAP followed by a 45 Gy-radiotherapy) in 1 patient. Among these 34 patients, 6 received adjuvant radiotherapy. 1 Supplemental Figure 2: Amino acid alignments of KIT H697 in the human protein and related orthologs, using (A) the Homologene database (exons 14 and 15), and (B) the UCSC Genome Browser database (exon 14). Residue H697 is highlighted with red boxes. Both alignments indicate that residue H697 is highly conserved. 2 Supplemental Figure 3: Direct comparison of the genomic profiles of thymic squamous cell carcinomas (n=7) and lung primary squamous cell carcinomas (n=6). (A) Unsupervised clustering analysis. Gains are indicated in red, and losses in green, by genomic position along the 22 chromosomes. (B) Genomic profiles and recurrent copy number alterations in thymic carcinomas and lung squamous cell carcinomas. Gains are indicated in red, and losses in blue. 3 Supplemental Methods Mutational profiling The exonic regions of interest (NCBI Human Genome Build 36.1) were broken into amplicons of 500 bp or less, and specific primers were designed using Primer 3 (on the World Wide Web for general users and for biologist programmers (see Supplemental Table 2) [1]. -

UC Irvine UC Irvine Electronic Theses and Dissertations
UC Irvine UC Irvine Electronic Theses and Dissertations Title Generation and application of a chimeric model to examine human microglia responses to amyloid pathology Permalink https://escholarship.org/uc/item/38g5s0sj Author Hasselmann, Jonathan Publication Date 2021 Supplemental Material https://escholarship.org/uc/item/38g5s0sj#supplemental Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California UNIVERSITY OF CALIFORNIA, IRVINE Generation and application of a chimeric model to examine human microglia responses to amyloid pathology DISSERTATION Submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY In Biological Sciences By Jonathan Hasselmann Dissertation Committee: Associate Professor Mathew Blurton-Jones, Chair Associate Professor Kim Green Professor Brian Cummings 2021 Portions of the Introduction and Chapter 2 © 2020 Wiley Periodicals, Inc. Chapter 1 and a portion of Chapter 2 © 2019 Elsevier Inc. All other materials © 2021 Jonathan Hasselmann DEDICATION To Nikki and Isla: During this journey, I have been fortunate enough to have the love and support of two of the strongest ladies I have ever met. I can say without a doubt that I would not have made it here without the two of you standing behind me. I love you both with all my heart and this achievement belongs to the two of you just as much as it belongs to me. Thank you for always having my back. ii TABLE OF CONTENTS LIST OF FIGURES .......................................................................................................................v -

Table S1. 103 Ferroptosis-Related Genes Retrieved from the Genecards
Table S1. 103 ferroptosis-related genes retrieved from the GeneCards. Gene Symbol Description Category GPX4 Glutathione Peroxidase 4 Protein Coding AIFM2 Apoptosis Inducing Factor Mitochondria Associated 2 Protein Coding TP53 Tumor Protein P53 Protein Coding ACSL4 Acyl-CoA Synthetase Long Chain Family Member 4 Protein Coding SLC7A11 Solute Carrier Family 7 Member 11 Protein Coding VDAC2 Voltage Dependent Anion Channel 2 Protein Coding VDAC3 Voltage Dependent Anion Channel 3 Protein Coding ATG5 Autophagy Related 5 Protein Coding ATG7 Autophagy Related 7 Protein Coding NCOA4 Nuclear Receptor Coactivator 4 Protein Coding HMOX1 Heme Oxygenase 1 Protein Coding SLC3A2 Solute Carrier Family 3 Member 2 Protein Coding ALOX15 Arachidonate 15-Lipoxygenase Protein Coding BECN1 Beclin 1 Protein Coding PRKAA1 Protein Kinase AMP-Activated Catalytic Subunit Alpha 1 Protein Coding SAT1 Spermidine/Spermine N1-Acetyltransferase 1 Protein Coding NF2 Neurofibromin 2 Protein Coding YAP1 Yes1 Associated Transcriptional Regulator Protein Coding FTH1 Ferritin Heavy Chain 1 Protein Coding TF Transferrin Protein Coding TFRC Transferrin Receptor Protein Coding FTL Ferritin Light Chain Protein Coding CYBB Cytochrome B-245 Beta Chain Protein Coding GSS Glutathione Synthetase Protein Coding CP Ceruloplasmin Protein Coding PRNP Prion Protein Protein Coding SLC11A2 Solute Carrier Family 11 Member 2 Protein Coding SLC40A1 Solute Carrier Family 40 Member 1 Protein Coding STEAP3 STEAP3 Metalloreductase Protein Coding ACSL1 Acyl-CoA Synthetase Long Chain Family Member 1 Protein -

Telencephalon Transcriptome Analysis of Chronically Stressed Adult Zebrafish
www.nature.com/scientificreports OPEN Telencephalon transcriptome analysis of chronically stressed adult zebrafsh Received: 22 June 2018 Victoria Huang, Anderson A. Butler & Farah D. Lubin Accepted: 13 December 2018 Chronic stress leads to disruptions in learning and memory processes. The efects of chronic stress Published: xx xx xxxx experience on the adult zebrafsh brain, particularly the memory associated telencephalon brain region, is unclear. The goal of this study was to identify gene expression changes in the adult zebrafsh brain triggered by chronic unpredictable stress. Transcriptome analysis of the telencephalon revealed 155 diferentially expressed genes. Of these genes, some are critical genes involved in learning and memory, such as cdk5 and chrna7, indicating efects of chronic unpredictable stress on zebrafsh memory. Interestingly, several genes were annotated in the Orange domain, which is an amino acid sequence present in eukaryotic DNA-binding transcription repressors. Furthermore, we identifed hsd11b2, a cortisol inactivating gene, as chronic stress-responsive in the whole zebrafsh brain. Collectively, these fndings suggest that memory associated gene expression changes in adult zebrafsh telencephalon are afected by chronic stress experience. Stress experience and anxiety afect physiology, behavior, and brain gene expression across many taxa. Stress response is a short-term adaptation to challenges in the environment, however, when prolonged, leads to det- rimental pathologies and learning and memory impairment1,2. Tese health and behavioral consequences are refected in gene expression changes in the brain, particularly in regions involved in cognitive processes such as the hippocampal region3,4. Like other vertebrates, zebrafsh (Danio rerio) also exhibit behavioral and gene expression responses to stress and anxiety that allow them to be models for cognitive disorders and dysfunction. -

Analysis of Gene Expression from Systemic Lupus Erythematosus Synovium Reveals Myeloid Cell‑Driven Pathogenesis of Lupus Arthritis Erika L
www.nature.com/scientificreports OPEN Analysis of gene expression from systemic lupus erythematosus synovium reveals myeloid cell‑driven pathogenesis of lupus arthritis Erika L. Hubbard1*, Michelle D. Catalina1,2, Sarah Heuer1,3, Prathyusha Bachali1, Robert Robl1, Nicholas S. Geraci1,4, Amrie C. Grammer1,5 & Peter E. Lipsky1,5 Arthritis is a common manifestation of systemic lupus erythematosus (SLE) yet understanding of the underlying pathogenic mechanisms remains incomplete. We, therefore, interrogated gene expression profles of SLE synovium to gain insight into the nature of lupus arthritis (LA), using osteoarthritis (OA) and rheumatoid arthritis (RA) as comparators. Knee synovia from SLE, OA, and RA patients were analyzed for diferentially expressed genes (DEGs) and also by Weighted Gene Co‑expression Network Analysis (WGCNA) to identify modules of highly co‑expressed genes. Genes upregulated and/or co‑expressed in LA revealed numerous immune/infammatory cells dominated by a myeloid phenotype, in which pathogenic macrophages, myeloid‑lineage cells, and their secreted products perpetuate infammation, whereas OA was characterized by fbroblasts and RA of lymphocytes. Genes governing trafcking of immune cells into the synovium by chemokines were identifed, but not in situ generation of germinal centers (GCs). Gene Set Variation Analysis (GSVA) confrmed activation of specifc immune cell types in LA. Numerous therapies were predicted to target LA, including TNF, NFκB, MAPK, and CDK inhibitors. Detailed gene expression analysis identifed a unique pattern of cellular components and physiologic pathways operative in LA, as well as drugs potentially able to target this common manifestation of SLE. Systemic lupus erythematosus (SLE) is a complex autoimmune disease in which loss of self-tolerance gives rise to pathogenic autoantibodies causing widespread infammation and tissue damage 1. -

Differential Gene Regulation in Fibroblasts in Co-Culture with Keratinocytes and Head and Neck SCC Cells
ANTICANCER RESEARCH 35: 3253-3266 (2015) Differential Gene Regulation in Fibroblasts in Co-culture with Keratinocytes and Head and Neck SCC Cells MALIN HAKELIUS1, DANIEL SAIEPOUR1, HANNA GÖRANSSON2, KRISTOFER RUBIN3, BENGT GERDIN1 and DANIEL NOWINSKI1 Departments of 1Surgical Sciences, Plastic Surgery and 3Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden; 2Array Facility, Department of Medical Sciences, Uppsala University, Uppsala, Sweden Abstract. Background: While carcinoma-associated growth. In cancers, this microenvironment, or tumor stroma, fibroblasts (CAFs) support tumorigenesis, normal tissue constitutes the backbone of the tumor and is essential for the fibroblasts suppress tumor progression. Mechanisms behind cohesiveness of the tumor tissue the tumor’s ability to thrive conversion of fibroblasts into a CAF phenotype are largely (1). This stroma has considerable similarities with that of unrevealed. Materials and Methods: Transwell co-cultures non-malignant repair processes that are characterized by with fibroblasts in collagen gels and squamous-cell activation of fibroblasts and neoformation of stromal tissue, carcinoma (SCC) cells or normal oral keratinocytes (NOKs) which has led to the concept of a tumor as a "wound that in inserts. Differences in fibroblast global gene expression never heals" (2). were analyzed using Affymetrix arrays and subsequent A fibroblast phenotype characterized by expression of functional annotation and cluster analysis, as well as gene alpha-smooth muscle actin (SMA), platelet-derived growth set enrichment analysis were performed. Results: There were factor receptor-beta (PDGFR-β) and the pericyte marker 52 up-regulated and 30 down-regulated transcript IDs neuron glial antigen 2 (NG2) is regarded as a key cell in the (>2-fold, p<0.05) in fibroblasts co-cultured with SCC tumor stroma (3). -

A Transcriptomics-Based Meta-Analysis Combined
medRxiv preprint doi: https://doi.org/10.1101/2020.04.16.20061515; this version posted April 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . 1 A transcriptomics-based meta-analysis combined with machine learning 2 approach identifies a secretory biomarker panel for diagnosis of pancreatic 3 adenocarcinoma 4 Indu Khatri1,3, Manoj K. Bhasin1,2* 5 6 Affiliations: 7 1Division of IMBIO, Department of Medicine, Beth Israel Lahey Health, Harvard Medical 8 School, Boston MA 02215 9 2Department of Pediatrics and Biomedical Informatics, Children Healthcare of Atlanta, Emory 10 School of Medicine, Atlanta, GA 30322 11 3Division of Immunohematology and Blood transfusion, Leiden University Medical Center, 12 Leiden, The Netherlands, 2333ZA 13 14 15 *Corresponding Author 16 Manoj K. Bhasin, PhD 17 E-mail: [email protected] 18 Keywords: biomarker, pancreatic cancer, secretory, transcriptome, validation 19 NOTE: This preprint reports new research that has not been certified1 by peer review and should not be used to guide clinical practice. medRxiv preprint doi: https://doi.org/10.1101/2020.04.16.20061515; this version posted April 22, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . 20 Abstract 21 Pancreatic ductal adenocarcinoma (PDAC) is largely incurable due to late diagnosis and absence 22 of markers that are concordant with expression in several sample sources (i.e.