Standardizing Gene Product Nomenclature—A Call To
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

1 Vitamins D2 and D3 Have Overlapping but Different Effects On
medRxiv preprint doi: https://doi.org/10.1101/2020.12.16.20247700; this version posted February 22, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY 4.0 International license . Vitamins D2 and D3 have overlapping but different effects on human gene expression revealed through analysis of blood transcriptomes in a randomised double-blind placebo-controlled food-fortification trial Louise R. Durrant,1, 2, Giselda Buccaa,b, 1,3, Andrew Heskethb, 1, Carla Möller-Leveta, Laura Tripkovica, Huihai Wua, Kathryn H. Harta, John C. Mathersc, Ruan M. Elliotta, Susan A. Lanham-Newa & Colin P. Smitha,b, 3* a Department of Nutritional Sciences, School of Biosciences and Medicine, Faculty of Health and Medical Sciences, University of Surrey, Guildford, UK b School of Pharmacy and Biomolecular Sciences, University of Brighton, Brighton, UK c Human Nutrition Research Centre, Population Health Sciences Institute, Newcastle University, UK 1 These authors contributed equally 2 Current address: Yakult UK Ltd., Anteros, Odyssey Business Park, West End Road, South Ruislip, HA4 6QQ; formerly known as Louise Wilson. 3 Current address: School of Pharmacy and Biomolecular Sciences, University of Brighton, Brighton, UK *For correspondence: Prof Colin P. Smith, School of Pharmacy and Biomolecular Sciences, University of Brighton, Huxley Building, Moulsecoomb, Brighton, BN2 4GJ, UK; Tel. +44 (0)1273 642048. Email: [email protected] Key words: Vitamin D3, vitamin D2, 25-hydroxyvitamin D, human transcriptome, immunity, ethnic differences, Covid-19 Abstract For the first time, we report the influence of vitamin D2 and vitamin D3 on genome- wide gene expression in whole blood from healthy women representing two ethnic groups, white European and South Asian. -

Biomarker Discovery for Chronic Liver Diseases by Multi-Omics
www.nature.com/scientificreports OPEN Biomarker discovery for chronic liver diseases by multi-omics – a preclinical case study Daniel Veyel1, Kathrin Wenger1, Andre Broermann2, Tom Bretschneider1, Andreas H. Luippold1, Bartlomiej Krawczyk1, Wolfgang Rist 1* & Eric Simon3* Nonalcoholic steatohepatitis (NASH) is a major cause of liver fbrosis with increasing prevalence worldwide. Currently there are no approved drugs available. The development of new therapies is difcult as diagnosis and staging requires biopsies. Consequently, predictive plasma biomarkers would be useful for drug development. Here we present a multi-omics approach to characterize the molecular pathophysiology and to identify new plasma biomarkers in a choline-defcient L-amino acid-defned diet rat NASH model. We analyzed liver samples by RNA-Seq and proteomics, revealing disease relevant signatures and a high correlation between mRNA and protein changes. Comparison to human data showed an overlap of infammatory, metabolic, and developmental pathways. Using proteomics analysis of plasma we identifed mainly secreted proteins that correlate with liver RNA and protein levels. We developed a multi-dimensional attribute ranking approach integrating multi-omics data with liver histology and prior knowledge uncovering known human markers, but also novel candidates. Using regression analysis, we show that the top-ranked markers were highly predictive for fbrosis in our model and hence can serve as preclinical plasma biomarkers. Our approach presented here illustrates the power of multi-omics analyses combined with plasma proteomics and is readily applicable to human biomarker discovery. Nonalcoholic fatty liver disease (NAFLD) is the major liver disease in western countries and is ofen associated with obesity, metabolic syndrome, or type 2 diabetes. -

Watsonjn2018.Pdf (1.780Mb)
UNIVERSITY OF CENTRAL OKLAHOMA Edmond, Oklahoma Department of Biology Investigating Differential Gene Expression in vivo of Cardiac Birth Defects in an Avian Model of Maternal Phenylketonuria A THESIS SUBMITTED TO THE GRADUATE FACULTY In partial fulfillment of the requirements For the degree of MASTER OF SCIENCE IN BIOLOGY By Jamie N. Watson Edmond, OK June 5, 2018 J. Watson/Dr. Nikki Seagraves ii J. Watson/Dr. Nikki Seagraves Acknowledgements It is difficult to articulate the amount of gratitude I have for the support and encouragement I have received throughout my master’s thesis. Many people have added value and support to my life during this time. I am thankful for the education, experience, and friendships I have gained at the University of Central Oklahoma. First, I would like to thank Dr. Nikki Seagraves for her mentorship and friendship. I lucked out when I met her. I have enjoyed working on this project and I am very thankful for her support. I would like thank Thomas Crane for his support and patience throughout my master’s degree. I would like to thank Dr. Shannon Conley for her continued mentorship and support. I would like to thank Liz Bullen and Dr. Eric Howard for their training and help on this project. I would like to thank Kristy Meyer for her friendship and help throughout graduate school. I would like to thank my committee members Dr. Robert Brennan and Dr. Lilian Chooback for their advisement on this project. Also, I would like to thank the biology faculty and staff. I would like to thank the Seagraves lab members: Jailene Canales, Kayley Pate, Mckayla Muse, Grace Thetford, Kody Harvey, Jordan Guffey, and Kayle Patatanian for their hard work and support. -

The Myotonic Dystrophy Locus
J Med Genet: first published as 10.1136/jmg.28.2.84 on 1 February 1991. Downloaded from 848 Med Genet 1991; 28: 84-88 Identification of new DNA markers close to the myotonic dystrophy locus J D Brook, H G Harley, K V Walsh, S A Rundle, M J Siciliano, P S Harper, D J Shaw Abstract dominant condition with a prevalence of 1 in 8000. The most useful markers for the prenatal diagnosis The DM gene is located on the long arm of of myotonic dystrophy (DM) are APOC2 and chromosome 19 within the interval 19q13.2-19q13.3. CKM, both of which map proximal to DM. In order Recently, a consensus order for markers in this region to produce other markers useful for DM, we have has emerged, which reads from centromere to telo- screened genomic DNA libraries constructed from mere: BCL3-APOC2-CKM-ERCCI-DM-D19SSO- cell line 20XP3542-1-4, which contains 20 to 30 Mb D19S22.25 On the proximal side of DM, CKM and of human material including APOC2 and CKM. ERCCI map to the same 300 kb NotI fragment.6 For Of 51 human clones identified, seven map to ERCCI few linkage data are available, as no con- chromosome 17, four to chromosome 8, and nine to ventional RFLPs have been documented. CKM chromosome 19, and the remaining 31 were excluded is closely linked to DM with a Omax of 0-02 (Zmax= from chromosome 19 but not localised further. 22 20) and a -1 lod support interval of 0 to 0 05. -
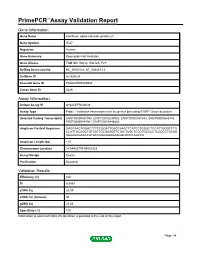
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name interferon, alpha-inducible protein 27 Gene Symbol IFI27 Organism Human Gene Summary Description Not Available Gene Aliases FAM14D, ISG12, ISG12A, P27 RefSeq Accession No. NC_000014.8, NT_026437.12 UniGene ID Hs.532634 Ensembl Gene ID ENSG00000165949 Entrez Gene ID 3429 Assay Information Unique Assay ID qHsaCEP0024638 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000557098, ENST00000298902, ENST00000557634, ENST00000555744, ENST00000444961, ENST00000448882 Amplicon Context Sequence GAGCAACTGGACTCTCCGGATTGACCAAGTTCATCCTGGGCTCCATTGGGTCTG CCATTGCGGCTGTCATTGCGAGGTTCTACTAGCTCCCTGCCCCTCGCCCTGCAG AGAAGAGAACCATGCCAGGGGAGAAGGCACCCAGCCA Amplicon Length (bp) 115 Chromosome Location 14:94582779-94582923 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 100 R2 0.9997 cDNA Cq 23.09 cDNA Tm (Celsius) 86 gDNA Cq 23.06 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report IFI27, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Human Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

Gene Nomenclature System for Rice
Rice (2008) 1:72–84 DOI 10.1007/s12284-008-9004-9 Gene Nomenclature System for Rice Susan R. McCouch & CGSNL (Committee on Gene Symbolization, Nomenclature and Linkage, Rice Genetics Cooperative) Received: 11 April 2008 /Accepted: 3 June 2008 /Published online: 15 August 2008 # The Author(s) 2008 Abstract The Committee on Gene Symbolization, Nomen- protein sequence information that have been annotated and clature and Linkage (CGSNL) of the Rice Genetics Cooper- mapped on the sequenced genome assemblies, as well as ative has revised the gene nomenclature system for rice those determined by biochemical characterization and/or (Oryza) to take advantage of the completion of the rice phenotype characterization by way of forward genetics. With genome sequence and the emergence of new methods for these revisions, we enhance the potential for structural, detecting, characterizing, and describing genes in the functional, and evolutionary comparisons across organisms biological community. This paper outlines a set of standard and seek to harmonize the rice gene nomenclature system procedures for describing genes based on DNA, RNA, and with that of other model organisms. Newly identified rice genes can now be registered on-line at http://shigen.lab.nig. ac.jp/rice/oryzabase_submission/gene_nomenclature/. The current ex-officio member list below is correct as of the date of this galley proof. The current ex-officio members of CGSNL (The Keywords Oryza sativa . Genome sequencing . Committee on Gene Symbolization, Nomenclature and Linkage) are: Gene symbolization Atsushi Yoshimura from the Faculty of Agriculture, Kyushu University, Fukuoka, 812-8581, Japan, email: [email protected] Guozhen Liu from the Beijing Institute of Genomics of the Chinese Introduction Academy of Sciences, Beijing, People’s Republic of China, email: [email protected] The biological community is moving towards a universal Yasuo Nagato from the Graduate School of Agricultural and Life system for the naming of genes. -

Down Regulation of Membrane-Bound Neu3 Constitutes a New
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Publications of the IAS Fellows IJC International Journal of Cancer Down regulation of membrane-bound Neu3 constitutes a new potential marker for childhood acute lymphoblastic leukemia and induces apoptosis suppression of neoplastic cells Chandan Mandal1, Cristina Tringali2, Susmita Mondal1, Luigi Anastasia2, Sarmila Chandra3, Bruno Venerando2 and Chitra Mandal1 1 Infectious Diseases and Immunology Division, Indian Institute of Chemical Biology, A Unit of Council of Scientific and Industrial Research, Govt of India, 4, Raja S. C. Mullick Road, Kolkata 700032, India 2 Department of Medical Chemistry, Biochemistry and Biotechnology, University of Milan, and IRCCS Policlinico San Donato, San Donato, Milan, Italy 3 Department of Hematology, Kothari Medical Centre, Kolkata 700027, India Membrane-linked sialidase Neu3 is a key enzyme for the extralysosomal catabolism of gangliosides. In this respect, it regulates pivotal cell surface events, including trans-membrane signaling, and plays an essential role in carcinogenesis. In this report, we demonstrated that acute lymphoblastic leukemia (ALL), lymphoblasts (primary cells from patients and cell lines) are characterized by a marked down-regulation of Neu3 in terms of both gene expression (230 to 40%) and enzymatic activity toward ganglioside GD1a (225.6 to 30.6%), when compared with cells from healthy controls. Induced overexpression of Neu3 in the ALL-cell line, MOLT-4, led to a significant increase of ceramide (166%) and to a parallel decrease of lactosylceramide (255%). These events strongly guided lymphoblasts to apoptosis, as we assessed by the decrease in Bcl2/ Bax ratio, the accumulation of Neu3 transfected cells in the sub G0–G1 phase of the cell cycle, the enhanced annexin-V positivity, the higher cleavage of procaspase-3. -

Evolution of Genomic Expression
C H A P T E R 5 Evolution of Genomic Expression Bernardo Lemos, Christian R. Landry, Pierre Fontanillas, Susan P. Renn, Rob Kulathinal, Kyle M. Brown, and Daniel L. Hartl Introduction Genomic regulation is key to cellular differentiation, tissue morphogenesis, and development. Increasing evidence indicates that evolutionary diversity of phenotypes—from cellular to organismic—may also be, in large part, the result of variation in the regulation of genomic expression. In this chapter we explore the complexity of gene regulation from the perspective of single genes and whole genomes. The first part describes the major factors affecting gene expression levels, from rates of gene transcrip- tion—as mediated by promoter–enhancer interactions and chromatin mod- ifications—to rates of mRNA degradation. This description underscores the multiple levels at which genomic expression can be regulated as well as the complexity and variety of mechanisms used. We then briefly describe the major experimental and computational biology techniques for analyzing gene expression variation and its underlying causes. The final section reviews our understanding of the role of regulatory variation in evolution, including the molecular evolution and population genetics of noncoding DNA, as well as the inheritance and phenotypic evolution of levels of mRNA abundance. The Complex Regulation of Genomic Expression The regulation of gene expression is a complex and dynamic process. It is not a simple matter to turn a gene on and off, let alone precisely regulate its level of expression. Regulation can be accomplished through various mech- anisms at nearly every step of the process of gene expression. Furthermore, each mechanism may require a variety of elements, including DNA sequences, RNA molecules, and proteins, acting in combination to deter- 2 Chapter Five Evolution of Genomic Expression 3 mine the final amount, timing, and location of functional gene product. -

Evidence for a Tumor Suppressor Gene on Chromosome 19Q Associated with Human Astrocytomas, Oligodendrogliomas, and Mixed Gliomas1
(CANCER RESEARCH 52. 4277-4279, August 1. 1992| Advances in Brief Evidence for a Tumor Suppressor Gene on Chromosome 19q Associated with Human Astrocytomas, Oligodendrogliomas, and Mixed Gliomas1 Andreas von Deimling,2 David N. Louis,3 Klaus von Ammon, Iver Petersen, Otmar D. Wiestier, and Bernd R. Seizinger Molecular Neuro-Oncology Laboratory and Neurosurgical Service, Massachusetts General Hospital and Harvard Medical School, Charlestown, Massachusetts 02129 [A. v. D., D. N. L,, B. R. S.J; Department of Pathology (Neuropathology), Massachusetts General Hospital and Harvard Medical School, Boston, Massachusetts 02114 ¡D.N. L.J; Department of Neurosurgery, University of Zürich,Raemistrasse 100, CH-8091 Zürich,Switzerland ¡K.v. A.]; and Laboratory of Neuropathology, Department of Pathology, University of Zurich, Schmelzbergstrasse 12, CH-8091 Zürich,Switzerland [L P., O. D. W.¡ Abstract is unique to fibrillary astrocytomas and Oligodendrogliomas, and whether a putative tumor suppressor gene locus exists on Previous studies have shown frequent allelic losses of chromosomes the long arm of chromosome 19, we evaluated 122 glial tumors 9p, 10, 17p, and 22q in glial tumors. Other researchers have briefly for LOH events on chromosome 19. reported that glial tumors may also show allelic losses of chromosome 19, suggesting a putative tumor suppressor gene locus on this chromo Materials and Methods some (D. T. Ransom et al., Proc. Am. Assoc. Cancer Res., 32: 302, 1991). To evaluate whether loss of chromosome 19 alÃelesis common in Tissue Specimens and Histopathology. Tumor and blood samples glial tumors of different types and grades, we performed Southern blot were obtained from 116 patients treated at the Massachusetts General restriction fragment length polymorphism analysis for multiple chro Hospital (Boston, MA) and at the University Hospital (Zurich, Swit mosome 19 loci in 122 gliomas from 116 patients. -

Age-Driven Developmental Drift in the Pathogenesis of Idiopathic Pulmonary Fibrosis
BACK TO BASICS INTERSTITIAL LUNG DISEASES | Age-driven developmental drift in the pathogenesis of idiopathic pulmonary fibrosis Moisés Selman1, Carlos López-Otín2 and Annie Pardo3 Affiliations: 1Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villegas, Mexico city, Mexico. 2Departamento de Bioquímica y Biología Molecular, Facultad de Medicina, Instituto Universitario de Oncología, Universidad de Oviedo, Oviedo, Spain. 3Facultad de Ciencias, Universidad Nacional Autónoma de México, Mexico city, Mexico. Correspondence: Moisés Selman, Instituto Nacional de Enfermedades Respiratorias, Tlalpan 4502, CP 14080, México DF, México. E-mail: [email protected] ABSTRACT Idiopathic pulmonary fibrosis (IPF) is a progressive and usually lethal disease of unknown aetiology. A growing body of evidence supports that IPF represents an epithelial-driven process characterised by aberrant epithelial cell behaviour, fibroblast/myofibroblast activation and excessive accumulation of extracellular matrix with the subsequent destruction of the lung architecture. The mechanisms involved in the abnormal hyper-activation of the epithelium are unclear, but we propose that recapitulation of pathways and processes critical to embryological development associated with a tissue specific age-related stochastic epigenetic drift may be implicated. These pathways may also contribute to the distinctive behaviour of IPF fibroblasts. Genomic and epigenomic studies have revealed that wingless/ Int, sonic hedgehog and other developmental signalling pathways are reactivated and deregulated in IPF. Moreover, some of these pathways cross-talk with transforming growth factor-β activating a profibrotic feedback loop. The expression pattern of microRNAs is also dysregulated in IPF and exhibits a similar expression profile to embryonic lungs. In addition, senescence, a process usually associated with ageing, which occurs early in alveolar epithelial cells of IPF lungs, likely represents a conserved programmed developmental mechanism. -

Regulation of Hematopoietic Activity Involving New Interacting Partners (RRAGC & PSMC2, CKAP4 & MANF and CTR9 & CNTNAP2)
CellBio, 2020, 9, 123-141 https://www.scirp.org/journal/cellbio ISSN Online: 2325-7792 ISSN Print: 2325-7776 Regulation of Hematopoietic Activity Involving New Interacting Partners (RRAGC & PSMC2, CKAP4 & MANF and CTR9 & CNTNAP2) Swati Sharma1, Gurudutta U. Gangenahalli2*, Upma Singh3 1Department of Pharmacology, All India Institute of Medical Science (AIIMS), New Delhi, India 2Stem Cell & Gene Therapy Research Group, Institute of Nuclear Medicine & Allied Sciences, New Delhi, India 3Department of Applied Chemistry, School of Vocational Studies & Applied Sciences, Gautam Buddha University, Greater Noida, India How to cite this paper: Sharma, S., Gan- Abstract genahalli, G.U. and Singh, U. (2020) Regu- lation of Hematopoietic Activity Involving Hematopoietic stem cells (HSCs) are tissue-specific cells giving rise to all New Interacting Partners (RRAGC & PSMC2, mature blood cell types regulated by a diverse group of hematopoietic cyto- CKAP4 & MANF and CTR9 & CNTNAP2). kines and growth factors that influences the survival & proliferation of early CellBio, 9, 123-141. https://doi.org/10.4236/cellbio.2020.93007 progenitors and differentiation mechanisms by modulating the functional ac- tivities of HSCs. In this study, the functional yet distinctive role of three novel Received: August 13, 2020 combinations of gene pairs RRAGC & PSMC2; CKAP4 & MANF; and CTR9 Accepted: September 27, 2020 & CNTNAP2 have been newly identified. These novel combinations of genes Published: September 30, 2020 were confirmed and expressed in K562 human leukemic cell line in the pres- Copyright © 2020 by author(s) and ence of cytokine combination (IL-3, FLT-3 and SCF) using RT-PCR and Scientific Research Publishing Inc. siRNA-mediated gene knock down strategy. -

INTRODUCTION Sirna and Rnai
J Korean Med Sci 2003; 18: 309-18 Copyright The Korean Academy ISSN 1011-8934 of Medical Sciences RNA interference (RNAi) is the sequence-specific gene silencing induced by dou- ble-stranded RNA (dsRNA). Being a highly specific and efficient knockdown tech- nique, RNAi not only provides a powerful tool for functional genomics but also holds Institute of Molecular Biology and Genetics and School of Biological Science, Seoul National a promise for gene therapy. The key player in RNAi is small RNA (~22-nt) termed University, Seoul, Korea siRNA. Small RNAs are involved not only in RNAi but also in basic cellular pro- cesses, such as developmental control and heterochromatin formation. The inter- Received : 19 May 2003 esting biology as well as the remarkable technical value has been drawing wide- Accepted : 23 May 2003 spread attention to this exciting new field. V. Narry Kim, D.Phil. Institute of Molecular Biology and Genetics and School of Biological Science, Seoul National University, San 56-1, Shillim-dong, Gwanak-gu, Seoul 151-742, Korea Key Words : RNA Interference (RNAi); RNA, Small interfering (siRNA); MicroRNAs (miRNA); Small Tel : +82.2-887-8734, Fax : +82.2-875-0907 hairpin RNA (shRNA); mRNA degradation; Translation; Functional genomics; Gene therapy E-mail : [email protected] INTRODUCTION established yet, testing 3-4 candidates are usually sufficient to find effective molecules. Technical expertise accumulated The RNA interference (RNAi) pathway was originally re- in the field of antisense oligonucleotide and ribozyme is now cognized in Caenorhabditis elegans as a response to double- being quickly applied to RNAi, rapidly improving RNAi stranded RNA (dsRNA) leading to sequence-specific gene techniques.