Telencephalon Transcriptome Analysis of Chronically Stressed Adult Zebrafish
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Variations in the PSMA6 and PSMC6 Proteasome Genes Are Associated with Multiple Sclerosis and Response to Interferon‑Β Therapy in Latvians
EXPERIMENTAL AND THERAPEUTIC MEDICINE 21: 478, 2021 Genetic variations in the PSMA6 and PSMC6 proteasome genes are associated with multiple sclerosis and response to interferon‑β therapy in Latvians NATALIA PARAMONOVA1, JOLANTA KALNINA1, KRISTINE DOKANE1, KRISTINE DISLERE1, ILVA TRAPINA1, TATJANA SJAKSTE1 and NIKOLAJS SJAKSTE1,2 1Genomics and Bioinformatics, Institute of Biology of The University of Latvia; 2Department of Medical Biochemistry of The University of Latvia, LV‑1004 Riga, Latvia Received July 8, 2020; Accepted December 8, 2020 DOI: 10.3892/etm.2021.9909 Abstract. Several polymorphisms in genes related to the Introduction ubiquitin‑proteasome system exhibit an association with pathogenesis and prognosis of various human autoimmune Multiple sclerosis (MS) is a lifelong demyelinating disease of diseases. Our previous study reported the association the central nervous system. The clinical onset of MS tends to between multiple sclerosis (MS) and the PSMA3‑rs2348071 be between the second and fourth decade of life. Similarly to polymorphism in the Latvian population. The current study other autoimmune diseases, women are affected 3‑4 times more aimed to evaluate the PSMA6 and PSMC6 genetic variations, frequently than men (1). About 10% of MS patients experience their interaction between each other and with the rs2348071, a primary progressive MS form characterized by the progres‑ on the susceptibility to MS risk and response to therapy in sion of neurological disability from the onset. In about 90% the Latvian population. PSMA6‑rs2277460, ‑rs1048990 and of MS patients, the disease undergoes the relapse‑remitting PSMC6‑rs2295826, ‑rs2295827 were genotyped in the MS MS course (RRMS); in most of these patients, the condition case/control study and analysed in terms of genotype‑protein acquires secondary progressive course (SPMS) (2). -

Identification of the Binding Partners for Hspb2 and Cryab Reveals
Brigham Young University BYU ScholarsArchive Theses and Dissertations 2013-12-12 Identification of the Binding arP tners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non- Redundant Roles for Small Heat Shock Proteins Kelsey Murphey Langston Brigham Young University - Provo Follow this and additional works at: https://scholarsarchive.byu.edu/etd Part of the Microbiology Commons BYU ScholarsArchive Citation Langston, Kelsey Murphey, "Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins" (2013). Theses and Dissertations. 3822. https://scholarsarchive.byu.edu/etd/3822 This Thesis is brought to you for free and open access by BYU ScholarsArchive. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston A thesis submitted to the faculty of Brigham Young University in partial fulfillment of the requirements for the degree of Master of Science Julianne H. Grose, Chair William R. McCleary Brian Poole Department of Microbiology and Molecular Biology Brigham Young University December 2013 Copyright © 2013 Kelsey Langston All Rights Reserved ABSTRACT Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactors and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston Department of Microbiology and Molecular Biology, BYU Master of Science Small Heat Shock Proteins (sHSP) are molecular chaperones that play protective roles in cell survival and have been shown to possess chaperone activity. -
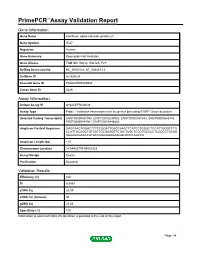
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name interferon, alpha-inducible protein 27 Gene Symbol IFI27 Organism Human Gene Summary Description Not Available Gene Aliases FAM14D, ISG12, ISG12A, P27 RefSeq Accession No. NC_000014.8, NT_026437.12 UniGene ID Hs.532634 Ensembl Gene ID ENSG00000165949 Entrez Gene ID 3429 Assay Information Unique Assay ID qHsaCEP0024638 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENST00000557098, ENST00000298902, ENST00000557634, ENST00000555744, ENST00000444961, ENST00000448882 Amplicon Context Sequence GAGCAACTGGACTCTCCGGATTGACCAAGTTCATCCTGGGCTCCATTGGGTCTG CCATTGCGGCTGTCATTGCGAGGTTCTACTAGCTCCCTGCCCCTCGCCCTGCAG AGAAGAGAACCATGCCAGGGGAGAAGGCACCCAGCCA Amplicon Length (bp) 115 Chromosome Location 14:94582779-94582923 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 100 R2 0.9997 cDNA Cq 23.09 cDNA Tm (Celsius) 86 gDNA Cq 23.06 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report IFI27, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Human Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

Regulation of NUB1 Activity Through Non-Proteolytic Mdm2-Mediated Ubiquitination
RESEARCH ARTICLE Regulation of NUB1 Activity through Non- Proteolytic Mdm2-Mediated Ubiquitination Thomas Bonacci, SteÂphane Audebert, Luc Camoin, Emilie Baudelet, Juan-Lucio Iovanna, Philippe Soubeyran* Centre de Recherche en CanceÂrologie de Marseille (CRCM), INSERM U1068, CNRS UMR 7258, Aix- Marseille Universite and Institut Paoli-Calmettes, Parc Scientifique et Technologique de Luminy, Marseille, France * [email protected] a1111111111 a1111111111 a1111111111 a1111111111 Abstract a1111111111 NUB1 (Nedd8 ultimate buster 1) is an adaptor protein which negatively regulates the ubiqui- tin-like protein Nedd8 as well as neddylated proteins levels through proteasomal degrada- tion. However, molecular mechanisms underlying this function are not completely understood. Here, we report that the oncogenic E3 ubiquitin ligase Mdm2 is a new NUB1 OPEN ACCESS interacting protein which induces its ubiquitination. Interestingly, we found that Mdm2-medi- Citation: Bonacci T, Audebert S, Camoin L, ated ubiquitination of NUB1 is not a proteolytic signal. Instead of promoting the conjugation Baudelet E, Iovanna J-L, Soubeyran P (2017) of polyubiquitin chains and the subsequent proteasomal degradation of NUB1, Mdm2 rather Regulation of NUB1 Activity through Non- Proteolytic Mdm2-Mediated Ubiquitination. PLoS induces its di-ubiquitination on lysine 159. Importantly, mutation of lysine 159 into arginine ONE 12(1): e0169988. doi:10.1371/journal. inhibits NUB1 activity by impairing its negative regulation of Nedd8 and of neddylated pro- pone.0169988 teins. We conclude that Mdm2 acts as a positive regulator of NUB1 function, by modulating Editor: Chunhong Yan, Augusta University, NUB1 ubiquitination on lysine 159. UNITED STATES Received: August 2, 2016 Accepted: December 27, 2016 Published: January 18, 2017 Introduction Copyright: © 2017 Bonacci et al. -

Modulating Antiviral Response Via CRISPR–Cas Systems
viruses Review Immunity and Viral Infections: Modulating Antiviral Response via CRISPR–Cas Systems Sergey Brezgin 1,2,3,† , Anastasiya Kostyusheva 1,†, Ekaterina Bayurova 4 , Elena Volchkova 5, Vladimir Gegechkori 6 , Ilya Gordeychuk 4,7, Dieter Glebe 8 , Dmitry Kostyushev 1,3,*,‡ and Vladimir Chulanov 1,3,5,‡ 1 National Medical Research Center of Tuberculosis and Infectious Diseases, Ministry of Health, 127994 Moscow, Russia; [email protected] (S.B.); [email protected] (A.K.); [email protected] (V.C.) 2 Institute of Immunology, Federal Medical Biological Agency, 115522 Moscow, Russia 3 Scientific Center for Genetics and Life Sciences, Division of Biotechnology, Sirius University of Science and Technology, 354340 Sochi, Russia 4 Chumakov Federal Scientific Center for Research and Development of Immune-and-Biological Products of Russian Academy of Sciences, 108819 Moscow, Russia; [email protected] (E.B.); [email protected] (I.G.) 5 Department of Infectious Diseases, Sechenov University, 119991 Moscow, Russia; [email protected] 6 Department of Pharmaceutical and Toxicological Chemistry, Sechenov University, 119991 Moscow, Russia; [email protected] 7 Department of Organization and Technology of Immunobiological Drugs, Sechenov University, 119991 Moscow, Russia 8 National Reference Center for Hepatitis B Viruses and Hepatitis D Viruses, Institute of Medical Virology, Justus Liebig University of Giessen, 35392 Giessen, Germany; [email protected] * Correspondence: [email protected] † Co-first authors. Citation: Brezgin, S.; Kostyusheva, ‡ Co-senior authors. A.; Bayurova, E.; Volchkova, E.; Gegechkori, V.; Gordeychuk, I.; Glebe, Abstract: Viral infections cause a variety of acute and chronic human diseases, sometimes resulting D.; Kostyushev, D.; Chulanov, V. Immunity and Viral Infections: in small local outbreaks, or in some cases spreading across the globe and leading to global pandemics. -

IFI27 May Predict and Evaluate the Severity of Respiratory Syncytial Virus
Gao et al. Hereditas (2021) 158:3 https://doi.org/10.1186/s41065-020-00167-5 RESEARCH Open Access IFI27 may predict and evaluate the severity of respiratory syncytial virus infection in preterm infants Junyan Gao1, Xueping Zhu2, Mingfu Wu1, Lijun Jiang1, Fudong Wang1 and Shan He2,3* Abstract Background: Preterm infants are a special population that vulnerable to respiratory syncytial virus (RSV) infection and the lower respiratory tract infections (LRTIs) caused by RSV could be severe and even life-threating. The purpose of the present study was to identify candidate genes of preterm infants who are susceptible to RSV infection and provide a new insight into the pathogenesis of RSV infection. Methods: Three datasets (GSE77087, GSE69606 and GSE41374) containing 183 blood samples of RSV infected patients and 33 blood samples of healthy controls from Gene Expression Omnibus (GEO) database were downloaded and the differentially expressed genes (DEGs) were screened out. The function and pathway enrichments were analyzed through Database for Annotation, Visualization and Integrated Discovery (DAVID) website. The protein-protein interaction (PPI) network for DEGs was constructed through Search Tool for the Retrieval of Interacting Genes (STRING). The module analysis was performed by Cytoscape software and hub genes were identified. Clinical verification was employed to verify the expression level of top five hub genes among 72 infants including 50 RSV infected patients and 22 non-RSV-infected patients hospitalized in our center. Further, the RSV infected infants with high-expression IFI27 and those with low-expression IFI27 were compared (defined as higher or lower than the median mRNA level). -

IFI27 May Predict and Evaluate the Severity of Respiratory Syncytial Virus Infection in Preterm Infants
IFI27 May Predict and Evaluate the Severity of Respiratory Syncytial Virus Infection in Preterm Infants Junyan Gao Aliated Hospital of Yangzhou University https://orcid.org/0000-0001-6678-6147 Xueping Zhu Children's Hospital of Soochow University Mingfu Wu Aliated Hospital of Yangzhou University Lijun Jiang Aliated Hospital of Yangzhou University Fudong Wang Aliated Hospital of Yangzhou University Shan He ( [email protected] ) Research Keywords: respiratory syncytial virus, preterm infants, differentially expressed genes, protein‐protein interaction, pathway enrichment, interferon alpha inducible protein 27 Posted Date: July 15th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-42880/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published on January 2nd, 2021. See the published version at https://doi.org/10.1186/s41065-020-00167-5. Page 1/21 Abstract Background: Preterm infants are a special population that vulnerable to respiratory syncytial virus (RSV) infection and the lower respiratory tract infection (LRTIs) caused by RSV could be severe and even life- threating. The purpose of the present study was to identify candidate genes of preterm infants who are susceptible to RSV infection and provide a new insight into the pathogenesis of RSV infection. Methods: Three datasets (GSE77087, GSE69606 and GSE41374) containing 183 blood samples of RSV infected patients and 33 blood samples of healthy controls from Gene Expression Omnibus (GEO) database were downloaded and the differentially expressed genes (DEGs) were screened out. The function and pathway enrichments were analyzed through Database for Annotation, Visualization and Integrated Discovery (DAVID) website. -

Plasma Cells in Vitro Generation of Long-Lived Human
Downloaded from http://www.jimmunol.org/ by guest on September 24, 2021 is online at: average * The Journal of Immunology , 32 of which you can access for free at: 2012; 189:5773-5785; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication November 2012; doi: 10.4049/jimmunol.1103720 http://www.jimmunol.org/content/189/12/5773 In Vitro Generation of Long-lived Human Plasma Cells Mario Cocco, Sophie Stephenson, Matthew A. Care, Darren Newton, Nicholas A. Barnes, Adam Davison, Andy Rawstron, David R. Westhead, Gina M. Doody and Reuben M. Tooze J Immunol cites 65 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2012/11/16/jimmunol.110372 0.DC1 This article http://www.jimmunol.org/content/189/12/5773.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 24, 2021. The Journal of Immunology In Vitro Generation of Long-lived Human Plasma Cells Mario Cocco,*,1 Sophie Stephenson,*,1 Matthew A. -

Cysteine‑Rich 61‑Associated Gene Expression Profile Alterations in Human Glioma Cells
MOLECULAR MEDICINE REPORTS 16: 5561-5567, 2017 Cysteine‑rich 61‑associated gene expression profile alterations in human glioma cells RUI WANG1, BO WEI2, JUN WEI3, YU TIAN2 and CHAO DU2 Departments of 1Radiology, 2Neurosurgery and 3Science and Education Section, China‑Japan Union Hospital of Jilin University, Changchun, Jilin 130033, P.R. China Received January 28, 2016; Accepted February 20, 2017 DOI: 10.3892/mmr.2017.7216 Abstract. The present study aimed to investigate gene be critical for maintaining the role of CYR61 during cancer expression profile alterations associated with cysteine‑rich 61 progression. (CYR61) expression in human glioma cells. The GSE29384 dataset, downloaded from the Gene Expression Omnibus, Introduction includes three LN229 human glioma cell samples expressing CYR61 induced by doxycycline (Dox group), and three Cysteine‑rich 61 (CYR61) is a secreted, cysteine‑rich, control samples not exposed to doxycycline (Nodox group). heparin‑binding protein (1) involved in a variety of cellular Differentially expressed genes (DEGs) between the Dox and functions including adhesion, migration and proliferation (2). Nodox groups were identified with cutoffs of |log2 fold change Previously, Xie et al (3) reported that CYR61 was overexpressed (FC)|>0.5 and P<0.05. Gene ontology and Kyoto Encyclopedia in 66 primary gliomas compared with healthy brain samples, of Genes and Genomes pathway enrichment analyses for and that CYR61 expression was significantly correlated with DEGs were performed. Protein‑protein interaction (PPI) tumor grade and patient survival (3). CYR61‑overexpressing network and module analyses were performed to identify glioma cells were observed to have an increased proliferation the most important genes. -

"The Genecards Suite: from Gene Data Mining to Disease Genome Sequence Analyses". In: Current Protocols in Bioinformat
The GeneCards Suite: From Gene Data UNIT 1.30 Mining to Disease Genome Sequence Analyses Gil Stelzer,1,5 Naomi Rosen,1,5 Inbar Plaschkes,1,2 Shahar Zimmerman,1 Michal Twik,1 Simon Fishilevich,1 Tsippi Iny Stein,1 Ron Nudel,1 Iris Lieder,2 Yaron Mazor,2 Sergey Kaplan,2 Dvir Dahary,2,4 David Warshawsky,3 Yaron Guan-Golan,3 Asher Kohn,3 Noa Rappaport,1 Marilyn Safran,1 and Doron Lancet1,6 1Department of Molecular Genetics, Weizmann Institute of Science, Rehovot, Israel 2LifeMap Sciences Ltd., Tel Aviv, Israel 3LifeMap Sciences Inc., Marshfield, Massachusetts 4Toldot Genetics Ltd., Hod Hasharon, Israel 5These authors contributed equally to the paper 6Corresponding author GeneCards, the human gene compendium, enables researchers to effectively navigate and inter-relate the wide universe of human genes, diseases, variants, proteins, cells, and biological pathways. Our recently launched Version 4 has a revamped infrastructure facilitating faster data updates, better-targeted data queries, and friendlier user experience. It also provides a stronger foundation for the GeneCards suite of companion databases and analysis tools. Improved data unification includes gene-disease links via MalaCards and merged biological pathways via PathCards, as well as drug information and proteome expression. VarElect, another suite member, is a phenotype prioritizer for next-generation sequencing, leveraging the GeneCards and MalaCards knowledgebase. It au- tomatically infers direct and indirect scored associations between hundreds or even thousands of variant-containing genes and disease phenotype terms. Var- Elect’s capabilities, either independently or within TGex, our comprehensive variant analysis pipeline, help prepare for the challenge of clinical projects that involve thousands of exome/genome NGS analyses. -

NUB1 Rabbit Pab
Leader in Biomolecular Solutions for Life Science NUB1 Rabbit pAb Catalog No.: A18665 Basic Information Background Catalog No. This gene encodes a protein that functions as a negative regulator of NEDD8, a A18665 ubiquitin-like protein that conjugates with cullin family members in order to regulate vital biological events. The protein encoded by this gene regulates the NEDD8 Observed MW conjugation system post-transcriptionally by recruiting NEDD8 and its conjugates to the 71kDa proteasome for degradation. This protein interacts with the product of the AIPL1 gene, which is associated with Leber congenital amaurosis, an inherited retinopathy, and Calculated MW mutations in that gene can abolish interaction with this protein, which may contribute to 69kDa/70kDa the pathogenesis. This protein is also known to accumulate in Lewy bodies in Parkinson's disease and dementia with Lewy bodies, and in glial cytoplasmic inclusions Category in multiple system atrophy, with this abnormal accumulation being specific to alpha- synucleinopathy lesions. Alternative splicing of this gene results in multiple transcript Primary antibody variants. Applications WB, IF Cross-Reactivity Human, Mouse, Rat Recommended Dilutions Immunogen Information WB 1:500 - 1:2000 Gene ID Swiss Prot 51667 Q9Y5A7 IF 1:50 - 1:200 Immunogen Recombinant protein of human IFT140. Synonyms NUB1; BS4; NUB1L; NYREN18; NEDD8 ultimate buster 1 Contact Product Information 400-999-6126 Source Isotype Purification Rabbit IgG Affinity purification [email protected] www.abclonal.com.cn Storage Store at -20℃. Avoid freeze / thaw cycles. Buffer: PBS with 0.02% sodium azide,50% glycerol,pH7.3. Validation Data Western blot analysis of extracts of various cell lines, using NUB1 Rabbit pAb (A18665) at 1:1000 dilution. -

Identification of Key Genes and Pathways in Pancreatic Cancer
G C A T T A C G G C A T genes Article Identification of Key Genes and Pathways in Pancreatic Cancer Gene Expression Profile by Integrative Analysis Wenzong Lu * , Ning Li and Fuyuan Liao Department of Biomedical Engineering, College of Electronic and Information Engineering, Xi’an Technological University, Xi’an 710021, China * Correspondence: [email protected]; Tel.: +86-29-86173358 Received: 6 July 2019; Accepted: 7 August 2019; Published: 13 August 2019 Abstract: Background: Pancreatic cancer is one of the malignant tumors that threaten human health. Methods: The gene expression profiles of GSE15471, GSE19650, GSE32676 and GSE71989 were downloaded from the gene expression omnibus database including pancreatic cancer and normal samples. The differentially expressed genes between the two types of samples were identified with the Limma package using R language. The gene ontology functional and pathway enrichment analyses of differentially-expressed genes were performed by the DAVID software followed by the construction of a protein–protein interaction network. Hub gene identification was performed by the plug-in cytoHubba in cytoscape software, and the reliability and survival analysis of hub genes was carried out in The Cancer Genome Atlas gene expression data. Results: The 138 differentially expressed genes were significantly enriched in biological processes including cell migration, cell adhesion and several pathways, mainly associated with extracellular matrix-receptor interaction and focal adhesion pathway in pancreatic cancer. The top hub genes, namely thrombospondin 1, DNA topoisomerase II alpha, syndecan 1, maternal embryonic leucine zipper kinase and proto-oncogene receptor tyrosine kinase Met were identified from the protein–protein interaction network.