Fungal Endophytes As Source to Combat Bacterial Infections
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
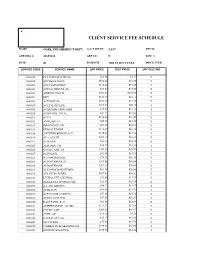
Oakland Sheriff's Dept Fee Schedule 02.09
CLIENT SERVICE FEE SCHEDULE NAME: OAKLAND SHERIFF'S DEPT ACCT SETUP: 2/1/87 REP ID: GRP BILL #: 22857610 GRP CD: N DISC %: FS ID: 01 FS DESCR: 2003 CLIENT FEES PRICE TYPE: SERVICE CODE SERVICE NAME UPF PRICE TEST PRICE UPF DISC IND 0000018 PLT SODIUM CITRATE $12.86 $4.37 Y 0000022 APC RESISTANCE $101.62 $34.55 Y 0000201 ACETAMINOPHEN $110.24 $37.48 Y 0000202 ACETALDEHYDE (B) $73.00 $73.00 N 0000203 ARSENIC,TISSUE $302.00 $302.00 N 0000204 DHT $182.70 $62.12 Y 0000205 ACETONE (B) $102.20 $34.75 Y 0000206 ACETYLCHOLINE $165.00 $56.10 Y 0000208 ACID PHOS, PROS, IMM $35.60 $12.10 Y 0000210 ACID PHOS, TOTAL $37.22 $12.65 Y 0000211 ACTH $110.00 $37.40 Y 0000212 AMYLASE (U) $45.45 $15.45 Y 0000213 IMMUNOFIX, UR $67.39 $22.91 Y 0000214 ETHOSUXIMIDE $112.07 $38.10 Y 0000216 ANTITHROMBIN III ACT $170.46 $57.96 Y 0000219 ALA, QUANT $107.10 $36.41 Y 0000223 ALBUMIN $22.88 $22.88 N 0000224 ALBUMIN, CSF $36.25 $12.33 Y 0000225 CYCLIC AMP, UR $205.80 $69.97 Y 0000227 ALDOLASE $32.08 $10.91 Y 0000228 A-2-MACROGLOB. $78.75 $26.78 Y 0000229 ALDOSTERONE (U) $251.58 $85.54 Y 0000230 ALDOSTERONE $207.10 $70.41 Y 0000231 ALK PHOS ISOENZYMES $61.34 $20.86 Y 0000232 AFP, FLUID W/RFX $107.40 $36.52 Y 0000233 LEUKOCYTE ALK PHOS $34.60 $11.76 Y 0000234 ALKALINE PHOSPHATASE $22.88 $22.88 N 0000235 A-1-ANTITRYPSN $40.22 $13.67 Y 0000236 AMIKACIN $99.91 $33.97 Y 0000237 AFP,TUMOR (CHIRON) $53.86 $18.31 Y 0000238 AMINO ACID SCR $87.15 $29.63 Y 0000240 RAST PANEL #103 $62.00 $62.00 N 0000241 AMPHETAMINE GC/MS $111.70 $37.98 Y 0000242 CYCLIC AMP $205.80 $69.97 Y 0000243 AMYLASE $16.42 $5.58 Y 0000248 HACKBERRY IGE $35.40 $35.40 N 0000249 ANA W/RFX $55.00 $18.70 Y 0000250 *AMIKACIN,PEAK&TROUGH $199.83 $67.94 Y 0000251 ANDROSTENEDIONE $180.65 $61.42 Y 0000252 ANTI-DIUR. -

Endophytic Fungi: Treasure for Anti-Cancerous Compounds
International Journal of Pharmacy and Pharmaceutical Sciences ISSN- 0975-1491 Vol 8, Issue 8, 2016 Review Article ENDOPHYTIC FUNGI: TREASURE FOR ANTI-CANCEROUS COMPOUNDS ANAND DILIP FIRODIYAa*, RAJESH KUMAR TENGURIAb aCSRD, Peoples University, Bhopal 462037, Madhya Pradesh, India, bDepartment of Botany, Govt. PG College, Rajgarh 496551, Madhya Pradesh, India Email: [email protected] Received: 22 Apr 2016 Revised and Accepted: 20 June 2016 ABSTRACT Endophytic fungi that live asymptomatically inside the plant tissues have novel bioactive metabolites exhibiting a variety of biological activities, especially against cancer. This review highlights the research progress on the production of anticancer compounds by endophytic fungi from 1990- 2015. Anticancer activity is generally associated with the cytotoxicity of the compounds present in the endophytic fungi. The ubiquitous nature of endophytic fungi synthesise diverse chemicals with promising anticancer activity from either their original host or related species. Modification in fermentation parameters and genetic insight of endophytes may produce novel anti-cancerous compounds. Keywords: Cancer, Medicinal plants, Secondary metabolites © 2016 The Authors. Published by Innovare Academic Sciences Pvt Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/) INTRODUCTION endophytic fungi detectable by high-performance liquid chromate- graphy, nuclear magnetic resonance, mass spectrophotometer and The interest in the biogenic medicines has revived throughout the X-ray crystallography and its cytotoxicity of the bioactive world, as the increase in awareness of the health hazards and compounds against cancer cell lines. The compounds with potential toxicity associated with the random use of synthetic drugs and application were also considered in the selection of antitumor antibiotics [1]. -

And Mushroom-Associated Alkaloids from Two Behavior Modifying
bioRxiv preprint doi: https://doi.org/10.1101/375105; this version posted December 18, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Psychoactive plant- and mushroom-associated alkaloids from two behavior modifying 2 cicada pathogens 3 4 Greg R. Boyce1, Emile Gluck-Thaler2, Jason C. Slot2, Jason E. Stajich3, William J. Davis4, Tim 5 Y. James4, John R. Cooley5, Daniel G. Panaccione1, Jørgen Eilenberg6, Henrik H. De Fine Licht6, 6 Angie M. Macias1, Matthew C. Berger1, Kristen L. Wickert1, Cameron M. Stauder1, Ellie J. 7 Spahr1, Matthew D. Maust1, Amy M. Metheny1, Chris Simon5, Gene Kritsky7, Kathie T. Hodge8, 8 Richard A. Humber8,9, Terry Gullion10, Dylan P. G. Short11, Teiya Kijimoto1, Dan Mozgai12, 9 Nidia Arguedas13, Matt T. Kasson1,* 10 11 1Division of Plant and Soil Sciences, West Virginia University, Morgantown, West Virginia, 12 26506, USA. 13 2Department of Plant Pathology, The Ohio State University, Columbus, Ohio 43210, USA. 14 3Department of Microbiology and Plant Pathology and Institute for Integrative Genome Biology, 15 University of California, Riverside, California 92521, USA. 16 4Department of Ecology and Evolution, University of Michigan, Ann Arbor, Michigan 48109, 17 USA. 18 5Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs, 19 Connecticut 06269, USA. 20 6Department of Plant and Environmental Science, University of Copenhagen, Denmark. 21 7Department of Biology, Mount St. Joseph University, Cincinnati, Ohio 45233, USA. 22 8Plant Pathology & Plant-Microbe Biology, School of Integrative Plant Science, Cornell 23 University, Ithaca, New York 14853, USA. -
Fungal Endophytes As Efficient Sources of Plant-Derived Bioactive
microorganisms Review Fungal Endophytes as Efficient Sources of Plant-Derived Bioactive Compounds and Their Prospective Applications in Natural Product Drug Discovery: Insights, Avenues, and Challenges Archana Singh 1,2, Dheeraj K. Singh 3,* , Ravindra N. Kharwar 2,* , James F. White 4,* and Surendra K. Gond 1,* 1 Department of Botany, MMV, Banaras Hindu University, Varanasi 221005, India; [email protected] 2 Department of Botany, Institute of Science, Banaras Hindu University, Varanasi 221005, India 3 Department of Botany, Harish Chandra Post Graduate College, Varanasi 221001, India 4 Department of Plant Biology, Rutgers University, New Brunswick, NJ 08901, USA * Correspondence: [email protected] (D.K.S.); [email protected] (R.N.K.); [email protected] (J.F.W.); [email protected] (S.K.G.) Abstract: Fungal endophytes are well-established sources of biologically active natural compounds with many producing pharmacologically valuable specific plant-derived products. This review details typical plant-derived medicinal compounds of several classes, including alkaloids, coumarins, flavonoids, glycosides, lignans, phenylpropanoids, quinones, saponins, terpenoids, and xanthones that are produced by endophytic fungi. This review covers the studies carried out since the first report of taxol biosynthesis by endophytic Taxomyces andreanae in 1993 up to mid-2020. The article also highlights the prospects of endophyte-dependent biosynthesis of such plant-derived pharma- cologically active compounds and the bottlenecks in the commercialization of this novel approach Citation: Singh, A.; Singh, D.K.; Kharwar, R.N.; White, J.F.; Gond, S.K. in the area of drug discovery. After recent updates in the field of ‘omics’ and ‘one strain many Fungal Endophytes as Efficient compounds’ (OSMAC) approach, fungal endophytes have emerged as strong unconventional source Sources of Plant-Derived Bioactive of such prized products. -

Fungal Endophytes: Promising Tools for Pharmaceutical Science
Int. J. Pharm. Sci. Rev. Res., 25(2), Mar – Apr 2014; Article No. 25, Pages: 128-138 ISSN 0976 – 044X Research Article Fungal Endophytes: Promising Tools for Pharmaceutical Science Pramod Kumar Pandey1,2, Siddhartha Singh1, Raj Naraian Singh Yadav2, Amit Kumar Singh1*, M. Chandra Kumar Singh1 1Department of Basic Science and Humanities, College of Horticulture and Forestry, Central Agricultural University, Pasighat, Arunachal Pradesh, India. 2Centre for Studies in Biotechnology, Dibrugarh University, Dibrugarh, Assam, India. *Corresponding author’s E-mail: [email protected] Accepted on: 27-01-2014; Finalized on: 31-03-2014. ABSTRACT Fungal endophytes are microorganisms that internally infect living plant tissues without causing any visible symptom of infection, and live in mutualistic relationship with plants for at least a part of their life cycle. Every plant in the world is reservoir of one or more number of endophytes. In recent year’s special attention have been made towards endophytic fungi because of their ability to synthesize several novel bioactive compounds not previously known to biological system which are important for pharmaceutical, agricultural and industrial sector. This review describes information on endophyte diversity, as well as production of secondary metabolites with special emphasis on anti cancerous, antimicrobial, antiviral, antibiotics along with huge number of other secondary metabolites for commercial exploitation in pharmaceutical and medical field. Furthermore, the chemical potential of endophytic fungi for drug discovery will be discussed with focus on the detection of pharmaceutically valuable plant constituents as products of fungal biosynthesis in recent years. At present a huge world population is suffering from the problem caused by drug resistant microbes (bacteria, parasitic protozoans and fungus) which decreases the efficiency of synthetic drugs. -

COMPARATIVE BIOLOGICAL EVALUATION of SIX ENDOPHYTIC FUNGI ISOLATED from VINCA ROSEA LEAVES Ahmed M
Az. J. Pharm Sci. Vol. 59, March, 2019. 137 COMPARATIVE BIOLOGICAL EVALUATION OF SIX ENDOPHYTIC FUNGI ISOLATED FROM VINCA ROSEA LEAVES Ahmed M. Metwaly*1, Mohamed A. Ashour1, Shabana Khan2, Guoyi Ma2, Hazem A. Kadry1, Atef A. El-Hela1, Abd-Elsalam I. Mohammad1, Stephen J. Cutler3 and Samir A. Ross2 1Department of Pharmacognosy, Faculty of Pharmacy, Al-Azhar University, Cairo, Egypt 2 Department of Biomolecular Science, National Center for Natural Products Research, University of Mississippi, Oxford, MS 38677, USA 3Department of Medicinal Chemistry, School of Pharmacy, the University of Mississippi, Oxford, MS 38677, USA *Corresponding authors: [email protected] Abstract In this study, a total of six endophytic fungi have been isolated from Vinca rosea (Apocynaceae) leaves growing in Egypt. The isolated fungi were identified morphologically and microscopically up to species to be; Alternaria phragmospora, Aspergillus awamori, Penicillium duclauxii, Penicillium melinii, Nigrospora sphaerica and Mucor ramosissimus. The extracts of the all identified fungi were screened biologically for antileukemic, cytotoxic, antimalarial, antileishmanial, antimicrobial, antioxidant and anti-inflammatory activities as well as for cannabinoid and opioid receptor binding affinities. Out of the six examined fungal extracts four exhibited promising antimalarial activities, four showed modeate antileukemic activities and two of them exhibited cytotoxic activities, three showed antioxidant activities, three exhibited anti-inflammatory activities, and the only one showed an antifungal activity. Alternaria phragmospora was the most active antimalarial agent inhibiting Plasmodium falciparum D6 and W2 clones with IC50 values of 1.9 and 2.1 μg/mL, respectively. Alternaria phragmospora extract was subjected to liquid-liquid partition using 90% MeOH, hexane then BuOH and H2O. -

Isolation and Characterization of Fungal Endophytes Isolated from Medicinal Plant Ephedra Pachyclada As Plant Growth-Promoting
biomolecules Article Isolation and Characterization of Fungal Endophytes Isolated from Medicinal Plant Ephedra pachyclada as Plant Growth-Promoting Ahmed Mohamed Aly Khalil 1,2, Saad El-Din Hassan 1,* , Sultan M. Alsharif 3, Ahmed M. Eid 1 , Emad El-Din Ewais 1, Ehab Azab 4,5 , Adil A. Gobouri 6, Amr Elkelish 7 and Amr Fouda 1,* 1 Department of Botany and Microbiology, Faculty of Science, Al-Azhar University, Nasr City, Cairo 11884, Egypt; [email protected] (A.M.A.K.); [email protected] (A.M.E.); [email protected] (E.E.-D.E.) 2 Biology Department, College of Science, Taibah University, Yanbu 41911, Saudi Arabia 3 Biology Department, Faculty of Science, Taibah University, Al Madinah P.O. Box 887, Saudi Arabia; [email protected] 4 Department of Biotechnology, College of Science, Taif University, P.O. Box 11099, Taif 21944, Saudi Arabia; [email protected] 5 Botany and Microbiology Department, Faculty of Science, Zagazig University, Zagazig 44519, Sharkia, Egypt 6 Department of Chemistry, College of Science, Taif University, P.O. Box 11099, Taif 21944, Saudi Arabia; [email protected] 7 Botany Department, Faculty of Science, Suez Canal University, Ismailia 41522, Egypt; [email protected] * Correspondence: [email protected] (S.E.-D.H.); [email protected] (A.F.); Citation: Khalil, A.M.A.; Tel.: +20-102-3884804 (S.E.-D.H.); +20-111-3351244 (A.F.) Hassan, S.E.-D.; Alsharif, S.M.; Eid, A.M.; Ewais, E.E.-D.; Azab, E.; Abstract: Endophytic fungi are widely present in internal plant tissues and provide different ben- Gobouri, A.A.; Elkelish, A.; Fouda, A. -

RSC COFI Prelims 1..4
The Chemistry of Fungi James R. Hanson Department of Chemistry, University of Sussex, Brighton, UK ISBN: 978-0-85404-136-7 A catalogue record for this book is available from the British Library r James R. Hanson, 2008 All rights reserved Apart from fair dealing for the purposes of research for non-commercial purposes or for private study, criticism or review, as permitted under the Copyright, Designs and Patents Act 1988 and the Copyright and Related Rights Regulations 2003, this publication may not be reproduced, stored or transmitted, in any form or by any means, without the prior permission in writing of The Royal Society of Chemistry or the copyright owner, or in the case of reproduction in accordance with the terms of licences issued by the Copyright Licensing Agency in the UK, or in accordance with the terms of the licences issued by the appropriate Reproduction Rights Organization outside the UK. Enquiries concerning reproduction outside the terms stated here should be sent to The Royal Society of Chemistry at the address printed on this page. Published by The Royal Society of Chemistry, Thomas Graham House, Science Park, Milton Road, Cambridge CB4 0WF, UK Registered Charity Number 207890 For further information see our web site at www.rsc.org Preface The diverse structures, biosyntheses and biological activities of fungal meta- bolites have attracted chemists for many years. This book is an introduction to the chemistry of fungal metabolites. The aim is to illustrate, within the context of fungal metabolites, the historical progression from chemical to spectroscopic methods of structure elucidation, the development in biosynthetic studies from establishing sequences and mechanisms to chemical enzymology and genetics and the increasing understanding of the biological roles of natural products. -

Laboratory & Diagnosis
Laboratory & Diagnosis Official Journal of Iranian Association of Clinical Laboratory Doctors Supplement Issue for IQC 17 Editorial Board Members: Dr. Kamaledine. Bagheri, DCLS Dr. Saeed Mahdavi, DCLS Dr. Mohammad Reza Bakhtiari, DCLS, PhD Dr. Ali Sadeghitabar, DCLS Dr. Mohammad Ghasem Eslami, DCLS Dr. Mohammad Sahebalzamani, DCLS Dr. S. Mohammad Hasan Hashemimadani, DCLS Dr. Narges Salajegheh, DCLS Executive Board Members: Ali Adibzadeh Sara Tondro Sedigheh Jalili Azam Jalili Mohammad Kazemi Leyla Pourdehghan Maryam Fazli Marzieh Moradi Mehrnosh shokrollahzadeh Layout by: Navid Ghahremani Circulation: 1000 copies Address: 1414734711, No.29, Ardeshir Alley, Hashtbehesht S t., Golha Square, Fatemi Ave, Tehran – Iran Telefax: (+98 21) 88970700 Laboratory & Diagnosis Vol. 10, No 42, Supplement Issue Message of Congress Chairman Dr. Mohammad Sahebalzamani DCLS In the Name of God We are planning to hold the 12th International Congress and 17th National Congress on Quality Improvement of Clinical Laboratory Services on 21-23 April 2019. We have tried to invite prominent national and international scientists and experts to take part in the event and present their scientific theories in order to ensure the improvement of congress’ quality. Congress authorities are going to provide the participants with a peaceful, friendly and jolly environment, together with scientific initiatives, as the outcome of scientific research of the congress. Modern researches of clinical sciences are in the course of rapid development and our country has a significant role in this venture. Hence, the authorities have to make a fundamental planning in order to increase enthusiasms and motivations of this large and educated s tratum. We mus t not isolate the scientific elements, scientists and skilled and experienced experts by performing incorrect activities and mus t avoid emptying out the scientific channels by superficial approaches. -

Chemical Diversity and Richness of Fungal Endophytes from Costa Rican Palicourea and Psychotria Species (Rubiaceae) 215-230 Acta Zoobot Austria 156, 2019, 215–230
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Verhandlungen der Zoologisch-Botanischen Gesellschaft in Wien. Frueher: Verh.des Zoologisch-Botanischen Vereins in Wien. seit 2014 "Acta ZooBot Austria" Jahr/Year: 2019 Band/Volume: 156 Autor(en)/Author(s): Hinterdobler Wolfgang, Schinnerl Johann Artikel/Article: Chemical diversity and richness of fungal endophytes from Costa Rican Palicourea and Psychotria species (Rubiaceae) 215-230 Acta ZooBot Austria 156, 2019, 215–230 Chemical diversity and richness of fungal endophytes from Costa Rican Palicourea and Psychotria species (Rubiaceae) Wolfgang Hinterdobler & Johann Schinnerl Screening of fungal endophytes from five Costa Rican Palicourea and three Psychotria species (Rubiaceae) growing in the surroundings of the Tropical Rainforest Station La Gamba resulted in the identification of strains belonging to the genera Xylaria, Arthrinium, Fusarium, Clonostachys and Colletotrichum. Metabolic profiles of isolated fungi were analyzed. Several cytochalasin derivatives, piliformic acid as well as the an- tifungal agents griseofulvin and its derivative 7-dechlorogriseofulvin were identified. Additionally, griseofulvin and its 7-dechloro form were found to be sequestered in the guttation droplets of four strains. Growth inhibiting effects against various microbial test organisms highlight the potential of the isolated fungi to produce powerful anti- biotic agents. HINTERDOBLER W. & SCHINNERL J., 2019: -

Antimicrobial Activity and GC-MS Analysis of Bioactive Constituents of Aspergillus Fumigatus 269 Isolated from Sungai Pinang Hot Spring, Riau, Indonesia
BIODIVERSITAS ISSN: 1412-033X Volume 22, Number 4, April 2021 E-ISSN: 2085-4722 Pages: 1839-1845 DOI: 10.13057/biodiv/d220429 Antimicrobial activity and GC-MS analysis of bioactive constituents of Aspergillus fumigatus 269 isolated from Sungai Pinang Hot Spring, Riau, Indonesia ZONA OCTARYA1,2, RIRYN NOVIANTY2, NABELLA SURAYA2, SARYONO2,♥ 1Department of Chemistry Education, Faculty of Tarbiyah and Teacher Training, Universitas Islam Negeri Sultan Syarif Kasim. Jl. Subrantas Km 15, Pekanbaru 28293, Riau, Indonesia 2Department of Chemistry, Faculty of Mathematics and Natural Science, Universitas Riau. Jl. Subrantas Km. 12,5, Kampus Bina Widya, Simpang Panam, Pekanbaru 28293, Riau, Indonesia. Tel./fax.: +62-761-63273, email: [email protected], [email protected] Manuscript received: 9 December 2020. Revision accepted: 20 March 2021. Abstract. Octarya Z, Novianty R, Suraya N, Saryono. 2021. Antimicrobial activity and GC-MS analysis of bioactive constituents of Aspergillus fumigatus 269 isolated from Sungai Pinang Hot Spring, Riau, Indonesia. Biodiversitas 22: 1839-1845. A total of 16 isolates of thermophilic fungi originating from hot springs in Riau and West Sumatra have been tested for their antimicrobial ability against pathogenic microbes Candida albicans, Staphylococcus aureus, and Escherichia coli. The antimicrobial test was carried out by using the disk diffusion method. Molecular identification of the most potential isolate (LBKURCC269) was carried out by amplifying the ITS (Internal transcribed spacer) sequence on rDNA using universal primer ITS-4 and ITS-5. ITS sequence results showed that LBKURCC269 has a 99% similarity to Aspergillus fumigatus. Ethyl acetate extract of LBKURCC269 (Aspergillus fumigatus 269) showed good antimicrobial activities against three pathogenic microbes tested with the inhibition of 17 mm, 13 mm, and 13 mm against Staphylococcus aureus, Candida albicans, and Escherichia coli, respectively. -

Secondary Metabolism Drives Ecological Breadth in the Xylariaceae 2 3 Mario E.E
bioRxiv preprint doi: https://doi.org/10.1101/2021.06.01.446356; this version posted June 2, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Secondary metabolism drives ecological breadth in the Xylariaceae 2 3 Mario E.E. Franco1, Jennifer H. Wisecaver2, A. Elizabeth Arnold3, Yu-Ming Ju4, Jason C. Slot5, Steven 4 Ahrendt6, Lillian P. Moore1, Katharine E. Eastman2, Kelsey Scott5, Zachary Konkel5, Stephen J. Mondo6, 5 Alan Kuo6, Richard Hayes6, Sajeet Haridas6, Bill Andreopoulos6, Robert Riley6, Kurt LaButti6, Jasmyn 6 Pangilinan6, Anna Lipzen6, Mojgan Amirebrahimi6, Juying Yan6, Catherine Adam6, Keykhosrow 7 Keymanesh6, Vivian Ng6, Katherine Louie6, Trent Northen6, Elodie Drula7, Bernard Henrissat7, Huei-Mei 8 Hsieh4, Ken Youens-Clark1, François Lutzoni8, Jolanta Miadlikowska8, Daniel C. Eastwood9, Richard C. 9 Hamelin10, Igor V. Grigoriev6,11, Jana M. U’Ren1* 10 11 12 13 1BIO5 Institute and Department of Biosystems Engineering, The University of Arizona, Tucson, AZ 14 85721 USA; 2Department of Biochemistry, Purdue University, West Lafayette, IN 47907 USA; 3School 15 of Plant Sciences and Department of Ecology and Evolutionary Biology, The University of Arizona, 16 Tucson, AZ 85721 USA; 4Institute of Plant and Microbial Biology, Academic Sinica, Taipei, Taiwan; 17 5Department of Plant Pathology, The Ohio State University, Columbus, OH, 43210