Title of Dissertation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
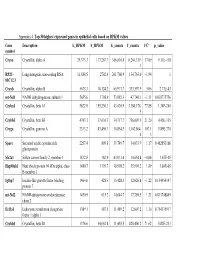
Appendix 4. Top 50 Highest Expressed Genes in Epithelial Cells Based on RPKM Values
Appendix 4. Top 50 highest expressed genes in epithelial cells based on RPKM values Gene Description E_RPKM F_RPKM E_counts F_counts FC* p_value symbol Cryaa Crystallin, alpha A 29,373.3 177,267.7 366,616.4 6,264,319. 17.09 9.11E-118 1 RP23– Long intergenic non-coding RNA 11,888.5 2702.4 261,760.9 134,763.0 −1.94 1 81C12.3 Cryab Crystallin, alpha B 5673.3 10,124.2 65,971.7 333,597.9 5.06 2.71E-43 mt-Nd1 NADH dehydrogenase, subunit 1 5655.6 1798.9 53,082.3 47,748.1 −1.11 0.838775756 Cryba1 Crystallin, beta A1 5622.0 155,230.3 43,420.9 3,380,176. 77.85 1.34E-240 5 Crybb3 Crystallin, beta B3 4743.1 37,636.3 34,717.7 736,007.9 21.20 4.45E-135 Cryga Crystallin, gamma A 2333.2 83,496.3 10,854.5 1,162,864. 107.1 5.89E-270 6 3 Sparc Secreted acidic cysteine rich 2257.4 809.8 39,749.7 34,033.9 −1.17 0.462853166 glycoprotein Slc2a1 Solute carrier family 2, member 1 1832.8 162.9 43,031.4 10,654.8 −4.04 1.67E-05 Hsp90ab1 Heat shock protein 90 kDa alpha, class 1480.7 1139.7 18,998.2 35,901.2 1.89 3.84E-05 B member 1 Igfbp7 Insulin-like growth factor binding 1464.6 428.3 15,428.3 12,626.8 −1.22 0.154954147 protein 7 mt-Nd2 NADH-ubiquinone oxidoreductase 1450.9 615.2 14,644.7 17,789.5 1.21 0.833748849 chain 2 Eef1a1 Eukaryotic translation elongation 1389.1 587.5 11,489.2 12,607.2 1.10 0.754135917 factor 1 alpha 1 Crybb1 Crystallin, beta B1 1376.6 34,662.8 11,455.5 820,406.2 71.62 5.82E-233 Htra3 HtrA serine peptidase 3 1338.6 162.0 23,197.6 6433.9 −3.61 3.93E-05 Gnb2l1 Guanine nucleotide-binding protein 1293.3 670.1 14,495.1 21,652.1 1.49 0.001685952 -

Supp Table 1.Pdf
Upregulated genes in Hdac8 null cranial neural crest cells fold change Gene Symbol Gene Title 134.39 Stmn4 stathmin-like 4 46.05 Lhx1 LIM homeobox protein 1 31.45 Lect2 leukocyte cell-derived chemotaxin 2 31.09 Zfp108 zinc finger protein 108 27.74 0710007G10Rik RIKEN cDNA 0710007G10 gene 26.31 1700019O17Rik RIKEN cDNA 1700019O17 gene 25.72 Cyb561 Cytochrome b-561 25.35 Tsc22d1 TSC22 domain family, member 1 25.27 4921513I08Rik RIKEN cDNA 4921513I08 gene 24.58 Ofa oncofetal antigen 24.47 B230112I24Rik RIKEN cDNA B230112I24 gene 23.86 Uty ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome 22.84 D8Ertd268e DNA segment, Chr 8, ERATO Doi 268, expressed 19.78 Dag1 Dystroglycan 1 19.74 Pkn1 protein kinase N1 18.64 Cts8 cathepsin 8 18.23 1500012D20Rik RIKEN cDNA 1500012D20 gene 18.09 Slc43a2 solute carrier family 43, member 2 17.17 Pcm1 Pericentriolar material 1 17.17 Prg2 proteoglycan 2, bone marrow 17.11 LOC671579 hypothetical protein LOC671579 17.11 Slco1a5 solute carrier organic anion transporter family, member 1a5 17.02 Fbxl7 F-box and leucine-rich repeat protein 7 17.02 Kcns2 K+ voltage-gated channel, subfamily S, 2 16.93 AW493845 Expressed sequence AW493845 16.12 1600014K23Rik RIKEN cDNA 1600014K23 gene 15.71 Cst8 cystatin 8 (cystatin-related epididymal spermatogenic) 15.68 4922502D21Rik RIKEN cDNA 4922502D21 gene 15.32 2810011L19Rik RIKEN cDNA 2810011L19 gene 15.08 Btbd9 BTB (POZ) domain containing 9 14.77 Hoxa11os homeo box A11, opposite strand transcript 14.74 Obp1a odorant binding protein Ia 14.72 ORF28 open reading -

A Comprehensive Analysis of the Expression of Crystallins in Mouse Retina Jinghua Xi Washington University School of Medicine in St
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2003 A comprehensive analysis of the expression of crystallins in mouse retina Jinghua Xi Washington University School of Medicine in St. Louis Rafal Farjo University of Michigan - Ann Arbor Shigeo Yoshida University of Michigan - Ann Arbor Timothy S. Kern Case Western Reserve University Anand Swaroop University of Michigan - Ann Arbor See next page for additional authors Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs Recommended Citation Xi, Jinghua; Farjo, Rafal; Yoshida, Shigeo; Kern, Timothy S.; Swaroop, Anand; and Andley, Usha P., ,"A comprehensive analysis of the expression of crystallins in mouse retina." Molecular Vision.9,. 410-419. (2003). https://digitalcommons.wustl.edu/open_access_pubs/1801 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. Authors Jinghua Xi, Rafal Farjo, Shigeo Yoshida, Timothy S. Kern, Anand Swaroop, and Usha P. Andley This open access publication is available at Digital Commons@Becker: https://digitalcommons.wustl.edu/open_access_pubs/1801 Molecular Vision 2003; 9:410-9 <http://www.molvis.org/molvis/v9/a53> © 2003 Molecular Vision Received 28 May 2003 | Accepted 19 August 2003 | Published 28 August 2003 A comprehensive analysis of the expression of crystallins in mouse retina Jinghua Xi,1 Rafal Farjo,3 Shigeo Yoshida,3 Timothy S. Kern,5 Anand Swaroop,3,4 Usha P. Andley1,2 Departments of 1Ophthalmology and Visual Sciences and 2Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. -

Omics in Myopia
Journal of Clinical Medicine Review Omics in Myopia Emil Tomasz Grochowski 1,* , Karolina Pietrowska 2 , Tomasz Kowalczyk 2, Zofia Mariak 1, Adam Kretowski 2,3, Michal Ciborowski 2,* and Diana Anna Dmuchowska 1,* 1 Department of Ophthalmology, Medical University of Bialystok, M. Sklodowskiej Curie 24a, 15-276 Bialystok, Poland; [email protected] 2 Clinical Research Centre, Medical University of Bialystok, M. Sklodowskiej Curie 24a, 15-276 Bialystok, Poland; [email protected] (K.P.); [email protected] (T.K.); [email protected] (A.K.) 3 Department of Endocrinology, Diabetology and Internal Medicine, Medical University of Bialystok, M. Sklodowskiej Curie 24a, 15-276 Bialystok, Poland * Correspondence: [email protected] (E.T.G.); [email protected] (M.C.); [email protected] (D.A.D.) Received: 8 October 2020; Accepted: 25 October 2020; Published: 28 October 2020 Abstract: Myopia is a globally emerging issue, with multiple medical and socio-economic burdens and no well-established causal treatment thus far. A better insight into altered biochemical pathways and underlying pathogenesis might facilitate early diagnosis and treatment of myopia, ultimately leading to the development of more effective preventive and therapeutic measures. In this review, we summarize current data about the metabolomics and proteomics of myopia in humans and present various experimental approaches and animal models, along with their strengths and weaknesses. We also discuss the potential applicability of these -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

Novel Mutation in the Γ-S Crystallin Gene Causing Autosomal Dominant Cataract
Molecular Vision 2009; 15:476-481 <http://www.molvis.org/molvis/v15/a48> © 2009 Molecular Vision Received 24 December 2008 | Accepted 27 February 2009 | Published 4 March 2009 Novel mutation in the γ-S crystallin gene causing autosomal dominant cataract Vanita Vanita,1 Jai Rup Singh,1 Daljit Singh,2 Raymonda Varon,3 Karl Sperling3 1Centre for Genetic Disorders, Guru Nanak Dev University, Amritsar, India; 2Dr. Daljit Singh Eye Hospital, Amritsar, India; 3Institute of Human Genetics, Charitè, University Medicine of Berlin, Berlin, Germany Purpose: To identify the underlying genetic defect in a north Indian family with seven members in three-generations affected with bilateral congenital cataract. Methods: Detailed family history and clinical data were recorded. Linkage analysis using fluorescently labeled microsatellite markers for the already known candidate gene loci was performed in combination with mutation screening by bidirectional sequencing. Results: Affected individuals had bilateral congenital cataract. Cataract was of opalescent type with the central nuclear region denser than the periphery. Linkage was excluded for the known cataract candidate gene loci at 1p34–36, 1q21–25 (gap junction protein, alpha 8 [GJA8]), 2q33–36 (crystallin, gamma A [CRYGA], crystallin, gamma B [CRYGB], crystallin, gamma C [CRYGC], crystallin, gamma D [CRYGD], crystallin, beta A2 [CRYBA2]), 3q21–22 (beaded filament structural protein 2, phakinin [BFSP2]), 12q12–14 (aquaporin 0 [AQP0]), 13q11–13 (gap junction protein, alpha 3 [GJA3]), 15q21– 22, 16q22–23 (v-maf musculoaponeurotic fibrosarcoma oncogene homolog [MAF], heat shock transcription factor 4 [HSF4]), 17q11–12 (crystallin, beta A1 [CRYBA1]), 17q24, 21q22.3 (crystallin, alpha A [CRYAA]), and 22q11.2 (crystallin, beta B1 [CRYBB1], crystallin, beta B2 [CRYBB2], crystallin, beta B3 [CRYBB3], crystallin, beta A4 [CRYBA4]). -

(12) Patent Application Publication (10) Pub. No.: US 2003/0198970 A1 Roberts (43) Pub
US 2003O19897OA1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2003/0198970 A1 Roberts (43) Pub. Date: Oct. 23, 2003 (54) GENOSTICS clinical trials on groups or cohorts of patients. This group data is used to derive a Standardised method of treatment (75) Inventor: Gareth Wyn Roberts, Cambs (GB) which is Subsequently applied on an individual basis. There is considerable evidence that a significant factor underlying Correspondence Address: the individual variability in response to disease, therapy and FINNEGAN, HENDERSON, FARABOW, prognosis lies in a person's genetic make-up. There have GARRETT & DUNNER been numerous examples relating that polymorphisms LLP within a given gene can alter the functionality of the protein 1300 ISTREET, NW encoded by that gene thus leading to a variable physiological WASHINGTON, DC 20005 (US) response. In order to bring about the integration of genomics into medical practice and enable design and building of a (73) Assignee: GENOSTIC PHARMA LIMITED technology platform which will enable the everyday practice (21) Appl. No.: 10/206,568 of molecular medicine a way must be invented for the DNA Sequence data to be aligned with the identification of genes (22) Filed: Jul. 29, 2002 central to the induction, development, progression and out come of disease or physiological States of interest. Accord Related U.S. Application Data ing to the invention, the number of genes and their configu rations (mutations and polymorphisms) needed to be (63) Continuation of application No. 09/325,123, filed on identified in order to provide critical clinical information Jun. 3, 1999, now abandoned. concerning individual prognosis is considerably less than the 100,000 thought to comprise the human genome. -

Mouse Models of Congenital Cataract
Mouse models of JOCHEN GRAW congenital cataract Abstract approach to collecting murine cataract mutations was initiated about 20 years ag02 Mouse mutants affecting lens development using paternal treatment of germ cells by X-raT are excellent models for corresponding human and later ethylnitrosourea.3 Among the disorders. The mutant aphakia has been offspring, dominant cataract mutations were characterised by bilaterally aphakic eyes identified by slit lamp observations and (Varnum and Stevens, J Hered 1968;59:147-50); subsequent genetic confirmation. The the corresponding gene was mapped to Neuherberg collection of cataracts contains now chromosome 19 (Varnum and Stevens, Mouse about 150 lines of independent origin and News Lett 1975;53:35). Recent investigations in distinct phenotypes.4,s our laboratory refined the linkage of 0.6 cM proximal to the marker D19Mitl0. Several candidate genes have been excluded (Chukl, Mouse models affecting early eye and lens FgfS, Lbpl, Npm3, Pax2, Pitx3). The Cat3 development mutations are characterised by vacuolated One of the most important genes in eye lenses caused by alterations in the initial development is the paired-box gene Pax6, which secondary lens fibre cell differentiation. is affected in various alleles of the mouse and Secondary malformations develop at the rat Small eye (Sey) mutants.6,7 In homozygous cornea and iris, but the retina remains Sey mice, eyes and nasal cavities do not unaffected. The mutation has been mapped to develop; the animals die soon after birth. The chromosome 10 close to the markers DlOMit41 histological analysis of homozygous mutants and DI0Mit95. Several candidate genes have demonstrated the presence of optic vesicles, but been excluded (Den, Elk3, Lde, MellS, Tr2-11). -

Bidirectional Analysis of Cryba4-Crybb1 Nascent Transcription and Nuclear Accumulation of Crybb3 Mrnas in Lens Fibers
Biochemistry and Molecular Biology Bidirectional Analysis of Cryba4-Crybb1 Nascent Transcription and Nuclear Accumulation of Crybb3 mRNAs in Lens Fibers Saima Limi,1 Yilin Zhao,1 Peng Guo,2 Melissa Lopez-Jones,2 Deyou Zheng,1,3,4 Robert H. Singer,2 Arthur I. Skoultchi,5 and Ales Cvekl1,6 1Departments of Genetics, Albert Einstein College of Medicine, Bronx, New York, United States 2Anatomy and Structural Biology, Albert Einstein College of Medicine, Bronx, New York, United States 3Neurology, Albert Einstein College of Medicine, Bronx, New York, United States 4Neuroscience, Albert Einstein College of Medicine, Bronx, New York, United States 5Cell Biology, Albert Einstein College of Medicine, Bronx, New York, United States 6Ophthalmology and Visual Sciences, Albert Einstein College of Medicine, Bronx, New York, United States Correspondence: Ales Cvekl, Oph- PURPOSE. Crystallin gene expression during lens fiber cell differentiation is tightly spatially and thalmology and Visual Sciences, Al- temporally regulated. A significant fraction of mammalian genes is transcribed from adjacent bert Einstein College of Medicine, promoters in opposite directions (‘‘bidirectional’’ promoters). It is not known whether two 1300 Morris Park Avenue, Ullmann proximal genes located on the same allele are simultaneously transcribed. 123, Bronx, New York, NY 10461, USA; METHODS. Mouse lens transcriptome was analyzed for paired genes whose transcriptional start [email protected]. sites are separated by less than 5 kbp to identify coexpressed bidirectional promoter gene Submitted: October 9, 2018 pairs. To probe these transcriptional mechanisms, nascent transcription of Cryba4, Crybb1, Accepted: November 26, 2018 and Crybb3 genes from gene-rich part of chromosome 5 was visualized by RNA fluorescent in situ hybridizations (RNA FISH) in individual lens fiber cell nuclei. -

A Novel Nonsense Mutation in CRYGC Is Associated with Autosomal Dominant Congenital Nuclear Cataracts and Microcornea
Molecular Vision 2009; 15:276-282 <http://www.molvis.org/molvis/v15/a28> © 2009 Molecular Vision Received 28 October 2008 | Accepted 29 January 2009 | Published 6 February 2009 A novel nonsense mutation in CRYGC is associated with autosomal dominant congenital nuclear cataracts and microcornea Lu Zhang,1 Songbin Fu,2 Yangshan Ou,1 Tingting Zhao,1 Yunjuan Su,1 Ping Liu1 1Eye hospital, the First Affiliated Hospital, Harbin Medical University, Harbin, China; 2Laboratory of Medical Genetics, Harbin Medical University, Harbin, China Purpose: To report the identification of a novel nonsense mutation in CRYGC in a Chinese family with autosomal dominant congenital nuclear cataracts and microcornea. Methods: We investigated a four-generation Chinese family with six members affected with nuclear cataracts and microcornea. The family resides in a relatively isolated region of northern China. Genomic DNA was isolated from blood leucocytes, genotyping was performed using more than 100 microsatellite markers for the known cataract candidate gene loci, and LOD scores were calculated using the LINKAGE programs. Mutations were detected by DNA sequence analysis of the candidate genes. Results: Evidence for linkage was detected at marker D2S325 (LOD score [Z]=2.29, recombination fraction [θ]=0.0), which closely flanks the γ-crystallin gene cluster (CRYGA-CRYGD) on chromosome 2q32.3-q35. Direct sequencing of the candidate CRYGA-CRYGD gene cluster revealed a c.470G>A transversion in exon 3 of CRYGC, which cosegregated with cataracts in the family and was not observed in 100 normal controls. This single nucleotide change was predicted to introduce a translation stop codon at tryptophan 157 (W157X). -

High-Throughput Genetic Screening of 51 Pediatric Cataract Genes Identifies Causative Mutations in Inherited Pediatric Cataract in South Eastern Australia
MUTANT SCREEN REPORT High-Throughput Genetic Screening of 51 Pediatric Cataract Genes Identifies Causative Mutations in Inherited Pediatric Cataract in South Eastern Australia Shari Javadiyan,*,1 Jamie E. Craig,* Emmanuelle Souzeau,* Shiwani Sharma,* Karen M. Lower,† David A. Mackey,‡,§,** Sandra E. Staffieri,§,**,†† James E. Elder,**,†† Deepa Taranath,* Tania Straga,‡‡ Joanna Black,‡‡ John Pater,‡‡ Theresa Casey,‡‡ Alex W. Hewitt,**,‡‡,§§,*** and Kathryn P. Burdon*,*** *Department of Ophthalmology and †Department of Haematology and Genetic Pathology, School of Medicine, Flinders University, Adelaide, South Australia 5042, Australia, ‡Centre for Ophthalmology and Visual Science, University of § Western Australia, Lions Eye Institute, Perth, Western Australia 6009, Australia, Centre for Eye Research Australia, Royal §§ Victorian Eye and Ear Hospital, Melbourne, Victoria 3002, Australia, **Department of Surgery and Department of Paediatrics, University of Melbourne, Victoria 3010, Australia, ††Department of Ophthalmology, Royal Children’sHospital, Melbourne, Victoria 3052, Australia, ‡‡Ophthalmology Department, Women’s and Children’s Hospital, Adelaide, South Australia 5006, Australia, and ***Menzies Institute for Medical Research, University of Tasmania, Hobart, Tasmania 7000, Australia ABSTRACT Pediatric cataract is a leading cause of childhood blindness. This study aimed to determine the KEYWORDS genetic cause of pediatric cataract in Australian families by screening known disease-associated genes using Ion Torrent massively parallel sequencing technology. We sequenced 51 previously reported pediatric cataract genes in PGM 33 affected individuals with a family history (cases with previously known or published mutations were excluded) congenital using the Ion Torrent Personal Genome Machine. Variants were prioritized for validation if they were predicted to cataract alter the protein sequence and were absent or rare with minor allele frequency ,1% in public databases. -

Lens Adaptation to Glutathione Deficiency
LENS ADAPTATION TO GLUTATHIONE DEFICIENCY: IMPLICATIONS FOR CATARACT by JEREMY A. WHITSON Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Department of Pathology CASE WESTERN RESERVE UNIVERSITY May, 2017 1 Table of Contents List of Tables 6 List of Figures 7 Acknowledgements 11 List of Abbreviations 12 Abstract 17 1. Introduction 19 1.1. The Crystalline Lens - Biological Glass 20 1.1.1. Anatomy of the Lens 20 1.1.2. Functions of the Lens 21 1.1.3. Lens Cell Biology 22 1.1.4. Transport and Homeostasis in the Lens 23 1.1.5. Conclusions 25 1.2. Age-Related Cataract - A Disease of Protein Aggregation 26 1.2.1. Pathology of Age-Related Cataract 26 1.2.2. Risk Factors for and Impact of Age-Related Cataract 27 1.2.3. Clinical Treatment of Cataract 28 1.2.4. Posterior Capsular Opacification 29 1.2.5. Conclusions 30 1.3. Glutathione - The Guardian of Lens Redox Homeostasis 30 1.3.1. Structure and Biosynthesis of Glutathione 30 1.3.2. Regulation of Cellular Glutathione Content 32 1.3.3. The Essential Role of Glutathione in the Lens 32 1.3.4. Lenticular Glutathione Declines with Age 34 1.3.5. A Loss of Glutathione Activity Leads to Cataract 34 1.3.6. The LEGSKO Mouse Model 35 1.3.7. A Salvage Pathway for Glutathione Synthesis 36 1.3.8. Conclusions 37 1.4. Glutathione Transport – Known Mechanisms 38 1.4.1. Glutathione Transport in the Lens 38 1.4.2.