Mutation G61C in the CRYGD Gene Causing Autosomal Dominant Congenital Coralliform Cataracts
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
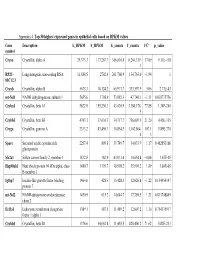
Appendix 4. Top 50 Highest Expressed Genes in Epithelial Cells Based on RPKM Values
Appendix 4. Top 50 highest expressed genes in epithelial cells based on RPKM values Gene Description E_RPKM F_RPKM E_counts F_counts FC* p_value symbol Cryaa Crystallin, alpha A 29,373.3 177,267.7 366,616.4 6,264,319. 17.09 9.11E-118 1 RP23– Long intergenic non-coding RNA 11,888.5 2702.4 261,760.9 134,763.0 −1.94 1 81C12.3 Cryab Crystallin, alpha B 5673.3 10,124.2 65,971.7 333,597.9 5.06 2.71E-43 mt-Nd1 NADH dehydrogenase, subunit 1 5655.6 1798.9 53,082.3 47,748.1 −1.11 0.838775756 Cryba1 Crystallin, beta A1 5622.0 155,230.3 43,420.9 3,380,176. 77.85 1.34E-240 5 Crybb3 Crystallin, beta B3 4743.1 37,636.3 34,717.7 736,007.9 21.20 4.45E-135 Cryga Crystallin, gamma A 2333.2 83,496.3 10,854.5 1,162,864. 107.1 5.89E-270 6 3 Sparc Secreted acidic cysteine rich 2257.4 809.8 39,749.7 34,033.9 −1.17 0.462853166 glycoprotein Slc2a1 Solute carrier family 2, member 1 1832.8 162.9 43,031.4 10,654.8 −4.04 1.67E-05 Hsp90ab1 Heat shock protein 90 kDa alpha, class 1480.7 1139.7 18,998.2 35,901.2 1.89 3.84E-05 B member 1 Igfbp7 Insulin-like growth factor binding 1464.6 428.3 15,428.3 12,626.8 −1.22 0.154954147 protein 7 mt-Nd2 NADH-ubiquinone oxidoreductase 1450.9 615.2 14,644.7 17,789.5 1.21 0.833748849 chain 2 Eef1a1 Eukaryotic translation elongation 1389.1 587.5 11,489.2 12,607.2 1.10 0.754135917 factor 1 alpha 1 Crybb1 Crystallin, beta B1 1376.6 34,662.8 11,455.5 820,406.2 71.62 5.82E-233 Htra3 HtrA serine peptidase 3 1338.6 162.0 23,197.6 6433.9 −3.61 3.93E-05 Gnb2l1 Guanine nucleotide-binding protein 1293.3 670.1 14,495.1 21,652.1 1.49 0.001685952 -

Supp Table 1.Pdf
Upregulated genes in Hdac8 null cranial neural crest cells fold change Gene Symbol Gene Title 134.39 Stmn4 stathmin-like 4 46.05 Lhx1 LIM homeobox protein 1 31.45 Lect2 leukocyte cell-derived chemotaxin 2 31.09 Zfp108 zinc finger protein 108 27.74 0710007G10Rik RIKEN cDNA 0710007G10 gene 26.31 1700019O17Rik RIKEN cDNA 1700019O17 gene 25.72 Cyb561 Cytochrome b-561 25.35 Tsc22d1 TSC22 domain family, member 1 25.27 4921513I08Rik RIKEN cDNA 4921513I08 gene 24.58 Ofa oncofetal antigen 24.47 B230112I24Rik RIKEN cDNA B230112I24 gene 23.86 Uty ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome 22.84 D8Ertd268e DNA segment, Chr 8, ERATO Doi 268, expressed 19.78 Dag1 Dystroglycan 1 19.74 Pkn1 protein kinase N1 18.64 Cts8 cathepsin 8 18.23 1500012D20Rik RIKEN cDNA 1500012D20 gene 18.09 Slc43a2 solute carrier family 43, member 2 17.17 Pcm1 Pericentriolar material 1 17.17 Prg2 proteoglycan 2, bone marrow 17.11 LOC671579 hypothetical protein LOC671579 17.11 Slco1a5 solute carrier organic anion transporter family, member 1a5 17.02 Fbxl7 F-box and leucine-rich repeat protein 7 17.02 Kcns2 K+ voltage-gated channel, subfamily S, 2 16.93 AW493845 Expressed sequence AW493845 16.12 1600014K23Rik RIKEN cDNA 1600014K23 gene 15.71 Cst8 cystatin 8 (cystatin-related epididymal spermatogenic) 15.68 4922502D21Rik RIKEN cDNA 4922502D21 gene 15.32 2810011L19Rik RIKEN cDNA 2810011L19 gene 15.08 Btbd9 BTB (POZ) domain containing 9 14.77 Hoxa11os homeo box A11, opposite strand transcript 14.74 Obp1a odorant binding protein Ia 14.72 ORF28 open reading -

A Comprehensive Analysis of the Expression of Crystallins in Mouse Retina Jinghua Xi Washington University School of Medicine in St
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2003 A comprehensive analysis of the expression of crystallins in mouse retina Jinghua Xi Washington University School of Medicine in St. Louis Rafal Farjo University of Michigan - Ann Arbor Shigeo Yoshida University of Michigan - Ann Arbor Timothy S. Kern Case Western Reserve University Anand Swaroop University of Michigan - Ann Arbor See next page for additional authors Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs Recommended Citation Xi, Jinghua; Farjo, Rafal; Yoshida, Shigeo; Kern, Timothy S.; Swaroop, Anand; and Andley, Usha P., ,"A comprehensive analysis of the expression of crystallins in mouse retina." Molecular Vision.9,. 410-419. (2003). https://digitalcommons.wustl.edu/open_access_pubs/1801 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. Authors Jinghua Xi, Rafal Farjo, Shigeo Yoshida, Timothy S. Kern, Anand Swaroop, and Usha P. Andley This open access publication is available at Digital Commons@Becker: https://digitalcommons.wustl.edu/open_access_pubs/1801 Molecular Vision 2003; 9:410-9 <http://www.molvis.org/molvis/v9/a53> © 2003 Molecular Vision Received 28 May 2003 | Accepted 19 August 2003 | Published 28 August 2003 A comprehensive analysis of the expression of crystallins in mouse retina Jinghua Xi,1 Rafal Farjo,3 Shigeo Yoshida,3 Timothy S. Kern,5 Anand Swaroop,3,4 Usha P. Andley1,2 Departments of 1Ophthalmology and Visual Sciences and 2Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. -

Related Macular Degeneration and Cutis Laxa
UvA-DARE (Digital Academic Repository) Genetic studies of age-related macular degeneration Baas, D.C. Publication date 2012 Document Version Final published version Link to publication Citation for published version (APA): Baas, D. C. (2012). Genetic studies of age-related macular degeneration. General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:05 Oct 2021 G������ S������ �� A��-������� M������ D����������� D����������� M������ G������ S������ �� A��-������� | 2012 D�������� C. B��� G������ S������ �� A��-������� M������ D����������� D�������� C. B��� cover.indd 1 31-10-12 08:36 Genetic Studies of Age-related Macular Degeneration Dominique C. Baas Chapter 0.indd 1 23-10-12 19:24 The research described in this thesis was conducted at the Netherlands Institute for Neuroscience (NIN), an institute of the Royal Netherlands Academy of Arts and Sciences, Department of Clinical and Molecular Ophthalmogenetics, Amsterdam, The Netherlands. -

Congenital Cataracts Due to a Novel 2‑Bp Deletion in CRYBA1/A3
1614 MOLECULAR MEDICINE REPORTS 10: 1614-1618, 2014 Congenital cataracts due to a novel 2‑bp deletion in CRYBA1/A3 JING ZHANG1, YANHUA ZHANG1, FANG FANG1, WEIHONG MU1, NING ZHANG2, TONGSHUN XU3 and QINYING CAO1 1Prenatal Diagnosis Center, Shijiazhuang Obstetrics and Gynecology Hospital; 2Department of Cardiology, The Second Hospital of Hebei Medical University; 3Department of Surgery, Shijiazhuang Obstetrics and Gynecology Hospital, Shijiazhuang, Hebei, P.R. China Received September 22, 2013; Accepted April 11, 2014 DOI: 10.3892/mmr.2014.2324 Abstract. Congenital cataracts, which are a clinically and located in the eye lens. The major human crystallins comprise genetically heterogeneous group of eye disorders, lead to 90% of protein in the mature lens and contain two different visual impairment and are a significant cause of blindness superfamilies: the small heat‑shock proteins (α-crystallins) in childhood. A major proportion of the causative mutations and the βγ-crystallins. for congenital cataracts are found in crystallin genes. In the In this study a functional candidate approach was used present study, a novel deletion mutation (c.590-591delAG) in to investigate the known crystallin genes, including CRYAA, exon 6 of CRYBA1/A3 was identified in a large family with CRYAB, CRYBA1/A3, CRYBB1, CRYBB2, CRYGC, CRYGD autosomal dominant congenital cataracts. An increase in and CRYGS, in which a major proportion of the mutations local hydrophobicity was predicted around the mutation site; identified in a large family with congenital cataracts were however, further studies are required to determine the exact found. effect of the mutation on βA1/A3-crystallin structure and function. To the best of our knowledge, this is the first report Subjects and methods of an association between a frameshift mutation in exon 6 of CRYBA1/A3 and congenital cataracts. -

Omics in Myopia
Journal of Clinical Medicine Review Omics in Myopia Emil Tomasz Grochowski 1,* , Karolina Pietrowska 2 , Tomasz Kowalczyk 2, Zofia Mariak 1, Adam Kretowski 2,3, Michal Ciborowski 2,* and Diana Anna Dmuchowska 1,* 1 Department of Ophthalmology, Medical University of Bialystok, M. Sklodowskiej Curie 24a, 15-276 Bialystok, Poland; [email protected] 2 Clinical Research Centre, Medical University of Bialystok, M. Sklodowskiej Curie 24a, 15-276 Bialystok, Poland; [email protected] (K.P.); [email protected] (T.K.); [email protected] (A.K.) 3 Department of Endocrinology, Diabetology and Internal Medicine, Medical University of Bialystok, M. Sklodowskiej Curie 24a, 15-276 Bialystok, Poland * Correspondence: [email protected] (E.T.G.); [email protected] (M.C.); [email protected] (D.A.D.) Received: 8 October 2020; Accepted: 25 October 2020; Published: 28 October 2020 Abstract: Myopia is a globally emerging issue, with multiple medical and socio-economic burdens and no well-established causal treatment thus far. A better insight into altered biochemical pathways and underlying pathogenesis might facilitate early diagnosis and treatment of myopia, ultimately leading to the development of more effective preventive and therapeutic measures. In this review, we summarize current data about the metabolomics and proteomics of myopia in humans and present various experimental approaches and animal models, along with their strengths and weaknesses. We also discuss the potential applicability of these -

A Novel Γd-Crystallin Mutation Causes Mild Changes in Protein Properties but Leads to Congenital Coralliform Cataract
Molecular Vision 2009; 15:1521-1529 <http://www.molvis.org/molvis/v15/a162> © 2009 Molecular Vision Received 29 April 2009 | Accepted 3 August 2009 | Published 6 August 2009 A novel γD-crystallin mutation causes mild changes in protein properties but leads to congenital coralliform cataract Li-Yun Zhang,1 Bo Gong,1 Jian-Ping Tong,2 Dorothy Shu-Ping Fan,1 Sylvia Wai-Yee Chiang,1 Dinghua Lou,2 Dennis Shun-Chiu Lam,1 Gary Hin-Fai Yam,1 Chi-Pui Pang1 1Department of Ophthalmology and Visual Sciences, The Chinese University of Hong Kong, Hong Kong, China; 2Department of Ophthalmology, the First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China Purpose: To identify the genetic lesions for congenital coralliform cataract. Methods: Two Chinese families with autosomal dominant coralliform cataract, 12 affected and 14 unaffected individuals, were recruited. Fifteen known genes associated with autosomal dominant congenital cataract were screened by two-point linkage analysis with gene based single nucleotide polymorphisms and microsatellite markers. Sequence variations were identified. Recombinant FLAG-tagged wild type or mutant γD-crystallin was expressed in human lens epithelial cells and COS-7 cells. Protein solubility and intracellular distribution were analyzed by western blotting and immunofluorescence, respectively. Results: A novel heterozygous change, c.43C>A (R15S) of γD-crystallin (CRYGD) co-segregated with coralliform cataract in one family and a known substitution, c.70C>A (P24T), in the other family. Unaffected family members and 103 unrelated control subjects did not carry these mutations. Similar to the wild type protein, R15S γD-crystallin was detergent soluble and was located in the cytoplasm. -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

A Recurrent Mutation in CRYGD Is Associated with Autosomal Dominant Congenital Coralliform Cataract in Two Unrelated Chinese Families
Molecular Vision 2011; 17:1085-1089 <http://www.molvis.org/molvis/v17/a123> © 2011 Molecular Vision Received 29 March 2011 | Accepted 19 April 2011 | Published 28 April 2011 A recurrent mutation in CRYGD is associated with autosomal dominant congenital coralliform cataract in two unrelated Chinese families Guoxing Yang,1 Chunlei Xiong,2 Shanlan Li,2 Yuanyuan Wang,2 Jialiang Zhao1 1Department of Opthalmology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China; 2Department of Opthalmology, Hai Cheng Rehabilitation Hospital, Liaoning, China Purpose: Congenital cataract is a clinically and genetically heterogeneous lens disorder. The purpose of this study was to identify the mutation responsible for autosomal dominant congenital coralliform cataracts in two Chinese families and to investigate the relationship between virulence genes and lens morphology. Methods: Patients received a physical examination, and blood samples were collected for DNA extraction. Mutation analysis was performed by direct sequencing of the candidate genes: gammaC-crystallin (CRYGC), gammaD-crystallin (CRYGD), gammaS-crystallin (CRYGS), gap-junction protein, alpha 8 (GJA8), gap-junction protein, alpha 3 (GJA3), and alphaA-crystallin (CRYAA). Results: The affected individuals in two families had congenital coralliform cataracts. Mutational analysis of the CRYGD identified a C→A transversion at nucleotide position c.70 in exon 2, which resulted in a threonine substitution for proline at amino-acid residue 24 (P24T). This mutation was identified in all affected individuals but was not found in healthy relatives or 100 control chromosomes from the same ethnic background. Conclusions: Our results indicated that the P24T mutation of CRYGD was responsible for two Chinese pedigrees with congenital coralliform cataracts. -

Crystallin, Γ-Crystallin and GJA8 Gene in Congenital Cataract
Molecular Vision 2011; 17:693-707 <http://www.molvis.org/molvis/v17/a79> © 2011 Molecular Vision Received 10 October 2010 | Accepted 7 March 2011 | Published 11 March 2011 Mutation screening and genotype phenotype correlation of α- crystallin, γ-crystallin and GJA8 gene in congenital cataract Manoj Kumar,1 Tushar Agarwal,2 Sudarshan Khokhar,2 Manoj Kumar,3 Punit Kaur,3 Tara Sankar Roy,4 Rima Dada1 1Laboratory for Molecular Reproduction and Genetics, Department of Anatomy, All India Institute of Medical Sciences, New Delhi, India; 2Dr. Rajendra Prasad Centre for Ophthalmic Sciences, All India Institute of Medical Sciences, New Delhi, India; 3Department of Biophysics, All India Institute of Medical Sciences, New Delhi, India; 4Department of Anatomy, All India Institute of Medical Sciences, New Delhi, India Purpose: To screen α-crystallin (CRYAB), γ-crystallin (CRYGC and CRYGD), and Connexin 50 (Cx-50 or GJA8) genes in congenital cataract patients and controls. Methods: Thirty clinically diagnosed congenital cataract cases below 3 years of age from northern India, presenting at Dr. R. P. Centre for Ophthalmic Sciences (AIIMS, New Delhi, India) were enrolled in this study. Genomic DNA was extracted from peripheral blood, all coding and exon/intron regions were amplified using PCR and direct sequencing was performed to detect any nucleotide variation. ProtScale and Discovery Studio programs were used for insilico and structural analysis of non-synonymous mutations. Results: DNA sequencing analysis of CRYAB, CRYGC, CRYGD, and GJA8 showed a total of six variations of which two were novel (CRYGC:p.R48H and GJA8:p.L281C) and four have been previously reported (CRYAB: rs11603779T>G, GJA8: p.L268L, CRYGD: p.R95R, and c.T564C). -

Zebrafish Model in Ophthalmology to Study Disease Mechanism And
pharmaceuticals Review Zebrafish Model in Ophthalmology to Study Disease Mechanism and Drug Discovery Yiwen Hong and Yan Luo * State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center, Sun Yat-Sen University, Guangzhou 510060, China; [email protected] * Correspondence: [email protected]; Tel.: +86-020-87335931 Abstract: Visual impairment and blindness are common and seriously affect people’s work and quality of life in the world. Therefore, the effective therapies for eye diseases are of high priority. Zebrafish (Danio rerio) is an alternative vertebrate model as a useful tool for the mechanism elucidation and drug discovery of various eye disorders, such as cataracts, glaucoma, diabetic retinopathy, age-related macular degeneration, photoreceptor degeneration, etc. The genetic and embryonic accessibility of zebrafish in combination with a behavioral assessment of visual function has made it a very popular model in ophthalmology. Zebrafish has also been widely used in ocular drug discovery, such as the screening of new anti-angiogenic compounds or neuroprotective drugs, and the oculotoxicity test. In this review, we summarized the applications of zebrafish as the models of eye disorders to study disease mechanism and investigate novel drug treatments. Keywords: zebrafish; eye; disease model; mechanism; drug candidate Citation: Hong, Y.; Luo, Y. Zebrafish Model in Ophthalmology to Study 1. Introduction Disease Mechanism and Drug Until 2020, an estimated 295 million people suffer from the moderate and severe visual Discovery. Pharmaceuticals 2021, 14, impairment worldwide, and among them, about 43.3 million people are even blind [1]. 716. https://doi.org/10.3390/ The leading global causes of blindness are cataract, followed by glaucoma, age-related ph14080716 macular degeneration (AMD), and diabetic retinopathy (DR) [2]. -

Rescue of Retinal Degeneration by Intravitreally Injected Adult Bone Marrow–Derived Lineage-Negative Hematopoietic Stem Cells
Rescue of retinal degeneration by intravitreally injected adult bone marrow–derived lineage-negative hematopoietic stem cells Atsushi Otani, … , John Heckenlively, Martin Friedlander J Clin Invest. 2004;114(6):765-774. https://doi.org/10.1172/JCI21686. Article Stem cells Inherited retinal degenerations afflict 1 in 3,500 individuals and are a heterogeneous group of diseases that result in profound vision loss, usually the result of retinal neuronal apoptosis. Atrophic changes in the retinal vasculature are also observed in many of these degenerations. While it is thought that this atrophy is secondary to diminished metabolic demand in the face of retinal degeneration, the precise relationship between the retinal neuronal and vascular degeneration is not clear. In this study we demonstrate that whenever a fraction of mouse or human adult bone marrow– derived stem cells (lineage-negative hematopoietic stem cells [Lin– HSCs]) containing endothelial precursors stabilizes and rescues retinal blood vessels that would ordinarily completely degenerate, a dramatic neurotrophic rescue effect is also observed. Retinal nuclear layers are preserved in 2 mouse models of retinal degeneration, rd1 and rd10, and detectable, albeit severely abnormal, electroretinogram recordings are observed in rescued mice at times when they are never observed in control-treated or untreated eyes. The normal mouse retina consists predominantly of rods, but the rescued cells after treatment with Lin– HSCs are nearly all cones. Microarray analysis of rescued retinas demonstrates