Supp Table 1.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

An Integrative Prognostic and Immune Analysis of PTPRD in Pan-Cancer
An Integrative Prognostic and Immune Analysis of PTPRD in Pan-Cancer Chunpei Ou longhua district central hospital Qin Peng longhua district central hospital Changchun Zeng ( [email protected] ) longhua district central hospital, guangdong medical university https://orcid.org/0000-0002-9489- 0627 Research Article Keywords: PTPRD, pan-cancer, prognosis, tumor-inltrating, immunotherapy Posted Date: June 4th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-569409/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/22 Abstract Background: PTPRD plays an indispensable role in the occurrence of multiple tumors. However, pan- cancer analysis is unavailable. The purpose of this research was to investigate the relationship between PTPRD and immunity and describe its prognostic landscape across various tumors. Methods: We explored expression prole, survival analysis, and genomic alterations of PTPRD based on the TIMER, GEPIA, UALCAN, PrognoScan, and cBioPortal database. The frequency of PTPRD mutation and its correlation with response to immunotherapy were evaluated using the cBioPortal database. The relationship between PTPRD and immune-cell inltration was analyzed by the TIMER and TISIDB databases. A protein interaction network was constructed by the STRING database. GO and KEGG enrichment analysis was executed by the Metascape database. Results: A signicant correlation between PTPRD expression and prognosis was found in various cancers. Aberrant PRPRD expression was closely related to immune inltration. Importantly, the patients who harbored PTPRD mutation and received immune checkpoint inhibitors had worse overall survival, especially in non-small cell lung cancer and melanoma, and had a higher TMB score. -

Supplemental Table S1
Entrez Gene Symbol Gene Name Affymetrix EST Glomchip SAGE Stanford Literature HPA confirmed Gene ID Profiling profiling Profiling Profiling array profiling confirmed 1 2 A2M alpha-2-macroglobulin 0 0 0 1 0 2 10347 ABCA7 ATP-binding cassette, sub-family A (ABC1), member 7 1 0 0 0 0 3 10350 ABCA9 ATP-binding cassette, sub-family A (ABC1), member 9 1 0 0 0 0 4 10057 ABCC5 ATP-binding cassette, sub-family C (CFTR/MRP), member 5 1 0 0 0 0 5 10060 ABCC9 ATP-binding cassette, sub-family C (CFTR/MRP), member 9 1 0 0 0 0 6 79575 ABHD8 abhydrolase domain containing 8 1 0 0 0 0 7 51225 ABI3 ABI gene family, member 3 1 0 1 0 0 8 29 ABR active BCR-related gene 1 0 0 0 0 9 25841 ABTB2 ankyrin repeat and BTB (POZ) domain containing 2 1 0 1 0 0 10 30 ACAA1 acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiol 0 1 0 0 0 11 43 ACHE acetylcholinesterase (Yt blood group) 1 0 0 0 0 12 58 ACTA1 actin, alpha 1, skeletal muscle 0 1 0 0 0 13 60 ACTB actin, beta 01000 1 14 71 ACTG1 actin, gamma 1 0 1 0 0 0 15 81 ACTN4 actinin, alpha 4 0 0 1 1 1 10700177 16 10096 ACTR3 ARP3 actin-related protein 3 homolog (yeast) 0 1 0 0 0 17 94 ACVRL1 activin A receptor type II-like 1 1 0 1 0 0 18 8038 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 1 0 0 0 0 19 8751 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 1 0 0 0 0 20 8728 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 1 0 0 0 0 21 81792 ADAMTS12 ADAM metallopeptidase with thrombospondin type 1 motif, 12 1 0 0 0 0 22 9507 ADAMTS4 ADAM metallopeptidase with thrombospondin type 1 -

Hemoglobin Interaction with Gp1ba Induces Platelet Activation And
ARTICLE Platelet Biology & its Disorders Hemoglobin interaction with GP1bα induces platelet activation and apoptosis: a novel mechanism associated with intravascular hemolysis Rashi Singhal,1,2,* Gowtham K. Annarapu,1,2,* Ankita Pandey,1 Sheetal Chawla,1 Amrita Ojha,1 Avinash Gupta,1 Miguel A. Cruz,3 Tulika Seth4 and Prasenjit Guchhait1 1Disease Biology Laboratory, Regional Centre for Biotechnology, National Capital Region, Biotech Science Cluster, Faridabad, India; 2Biotechnology Department, Manipal University, Manipal, Karnataka, India; 3Thrombosis Research Division, Baylor College of Medicine, Houston, TX, USA, and 4Hematology, All India Institute of Medical Sciences, New Delhi, India *RS and GKA contributed equally to this work. ABSTRACT Intravascular hemolysis increases the risk of hypercoagulation and thrombosis in hemolytic disorders. Our study shows a novel mechanism by which extracellular hemoglobin directly affects platelet activation. The binding of Hb to glycoprotein1bα activates platelets. Lower concentrations of Hb (0.37-3 mM) significantly increase the phos- phorylation of signaling adapter proteins, such as Lyn, PI3K, AKT, and ERK, and promote platelet aggregation in vitro. Higher concentrations of Hb (3-6 mM) activate the pro-apoptotic proteins Bak, Bax, cytochrome c, caspase-9 and caspase-3, and increase platelet clot formation. Increased plasma Hb activates platelets and promotes their apoptosis, and plays a crucial role in the pathogenesis of aggregation and development of the procoagulant state in hemolytic disorders. Furthermore, we show that in patients with paroxysmal nocturnal hemoglobinuria, a chronic hemolytic disease characterized by recurrent events of intravascular thrombosis and thromboembolism, it is the elevated plasma Hb or platelet surface bound Hb that positively correlates with platelet activation. -
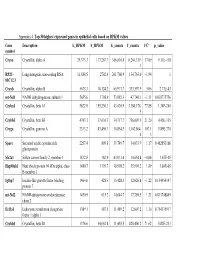
Appendix 4. Top 50 Highest Expressed Genes in Epithelial Cells Based on RPKM Values
Appendix 4. Top 50 highest expressed genes in epithelial cells based on RPKM values Gene Description E_RPKM F_RPKM E_counts F_counts FC* p_value symbol Cryaa Crystallin, alpha A 29,373.3 177,267.7 366,616.4 6,264,319. 17.09 9.11E-118 1 RP23– Long intergenic non-coding RNA 11,888.5 2702.4 261,760.9 134,763.0 −1.94 1 81C12.3 Cryab Crystallin, alpha B 5673.3 10,124.2 65,971.7 333,597.9 5.06 2.71E-43 mt-Nd1 NADH dehydrogenase, subunit 1 5655.6 1798.9 53,082.3 47,748.1 −1.11 0.838775756 Cryba1 Crystallin, beta A1 5622.0 155,230.3 43,420.9 3,380,176. 77.85 1.34E-240 5 Crybb3 Crystallin, beta B3 4743.1 37,636.3 34,717.7 736,007.9 21.20 4.45E-135 Cryga Crystallin, gamma A 2333.2 83,496.3 10,854.5 1,162,864. 107.1 5.89E-270 6 3 Sparc Secreted acidic cysteine rich 2257.4 809.8 39,749.7 34,033.9 −1.17 0.462853166 glycoprotein Slc2a1 Solute carrier family 2, member 1 1832.8 162.9 43,031.4 10,654.8 −4.04 1.67E-05 Hsp90ab1 Heat shock protein 90 kDa alpha, class 1480.7 1139.7 18,998.2 35,901.2 1.89 3.84E-05 B member 1 Igfbp7 Insulin-like growth factor binding 1464.6 428.3 15,428.3 12,626.8 −1.22 0.154954147 protein 7 mt-Nd2 NADH-ubiquinone oxidoreductase 1450.9 615.2 14,644.7 17,789.5 1.21 0.833748849 chain 2 Eef1a1 Eukaryotic translation elongation 1389.1 587.5 11,489.2 12,607.2 1.10 0.754135917 factor 1 alpha 1 Crybb1 Crystallin, beta B1 1376.6 34,662.8 11,455.5 820,406.2 71.62 5.82E-233 Htra3 HtrA serine peptidase 3 1338.6 162.0 23,197.6 6433.9 −3.61 3.93E-05 Gnb2l1 Guanine nucleotide-binding protein 1293.3 670.1 14,495.1 21,652.1 1.49 0.001685952 -

ANKRD11 Gene Ankyrin Repeat Domain 11
ANKRD11 gene ankyrin repeat domain 11 Normal Function The ANKRD11 gene provides instructions for making a protein called ankyrin repeat domain 11 (ANKRD11). As its name suggests, this protein contains multiple regions called ankyrin domains; proteins with these domains help other proteins interact with each other. The ANKRD11 protein interacts with certain proteins called histone deacetylases, which are important for controlling gene activity. Through these interactions, ANKRD11 affects when genes are turned on and off. For example, ANKRD11 brings together histone deacetylases and other proteins called p160 coactivators. This association regulates the ability of p160 coactivators to turn on gene activity. ANKRD11 may also enhance the activity of a protein called p53, which controls the growth and division (proliferation) and the self-destruction (apoptosis) of cells. The ANKRD11 protein is found in nerve cells (neurons) in the brain. During embryonic development, ANKRD11 helps regulate the proliferation of these cells and development of the brain. Researchers speculate that the protein may also be involved in the ability of neurons to change and adapt over time (plasticity), which is important for learning and memory. ANKRD11 may function in other cells in the body and appears to be involved in normal bone development. Health Conditions Related to Genetic Changes KBG syndrome Several ANKRD11 gene mutations have been found to cause KBG syndrome, a condition characterized by large upper front teeth and other unusual facial features, skeletal abnormalities, and intellectual disability. Most of these mutations lead to an abnormally short ANKRD11 protein, which likely has little or no function. Reduction of this protein's function is thought to underlie the signs and symptoms of the condition. -

Identification of Chebulinic Acid As a Dual Targeting Inhibitor of Protein
Bioorganic Chemistry 90 (2019) 103087 Contents lists available at ScienceDirect Bioorganic Chemistry journal homepage: www.elsevier.com/locate/bioorg Short communication Identification of chebulinic acid as a dual targeting inhibitor of protein T tyrosine phosphatases relevant to insulin resistance Sun-Young Yoona,1, Hyo Jin Kangb,1, Dohee Ahna, Ji Young Hwanga, Se Jeong Kwona, ⁎ Sang J. Chunga, a School of Pharmacy, Sungkyunkwan University, Suwon 16419, Republic of Korea b Department of Chemistry, Dongguk University, Seoul 100-715, Republic of Korea ARTICLE INFO ABSTRACT Keywords: Natural products as antidiabetic agents have been shown to stimulate insulin signaling via the inhibition of the Protein tyrosine phosphatases (PTPs) protein tyrosine phosphatases relevant to insulin resistance. Previously, we have identified PTPN9 and DUSP9 as Chebulinic acid potential antidiabetic targets and a multi-targeting natural product thereof. In this study, knockdown of PTPN11 Type 2 diabetes increased AMPK phosphorylation in differentiated C2C12 muscle cells by 3.8 fold, indicating that PTPN11 could Glucose-uptake be an antidiabetic target. Screening of a library of 658 natural products against PTPN9, DUSP9, or PTPN11 PTPN9 identified chebulinic acid (CA) as a strong allosteric inhibitor with a slow cooperative binding toPTPN9 PTPN11 (IC50 = 34 nM) and PTPN11 (IC50 = 37 nM), suggesting that it would be a potential antidiabetic candidate. Furthermore, CA stimulated glucose uptake and resulted in increased AMP-activated protein kinase (AMPK) phosphorylation. Taken together, we demonstrated that CA increased glucose uptake as a dual inhibitor of PTPN9 and PTPN11 through activation of the AMPK signaling pathway. These results strongly suggest that CA could be used as a potential therapeutic candidate for the treatment of type 2 diabetes. -

Meconium Ileus Caused by Mutations in GUCY2C, Encoding the CFTR-Activating Guanylate Cyclase 2C
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector REPORT Meconium Ileus Caused by Mutations in GUCY2C, Encoding the CFTR-Activating Guanylate Cyclase 2C Hila Romi,1,6 Idan Cohen,1,6 Daniella Landau,2 Suliman Alkrinawi,2 Baruch Yerushalmi,2 Reli Hershkovitz,3 Nitza Newman-Heiman,2 Garry R. Cutting,4 Rivka Ofir,1 Sara Sivan,1 and Ohad S. Birk1,5,* Meconium ileus, intestinal obstruction in the newborn, is caused in most cases by CFTR mutations modulated by yet-unidentified modi- fier genes. We now show that in two unrelated consanguineous Bedouin kindreds, an autosomal-recessive phenotype of meconium ileus that is not associated with cystic fibrosis (CF) is caused by different homozygous mutations in GUCY2C, leading to a dramatic reduction or fully abrogating the enzymatic activity of the encoded guanlyl cyclase 2C. GUCY2C is a transmembrane receptor whose extracellular domain is activated by either the endogenous ligands, guanylin and related peptide uroguanylin, or by an external ligand, Escherichia coli (E. coli) heat-stable enterotoxin STa. GUCY2C is expressed in the human intestine, and the encoded protein activates the CFTR protein through local generation of cGMP. Thus, GUCY2C is a likely candidate modifier of the meconium ileus phenotype in CF. Because GUCY2C heterozygous and homozygous mutant mice are resistant to E. coli STa enterotoxin-induced diarrhea, it is plausible that GUCY2C mutations in the desert-dwelling Bedouin kindred are of selective advantage. Meconium ileus (MI), intestinal obstruction by inspissated homozygosity on chromosome 12p13 (spanning 9.5 Mb meconium in the distal ileum and cecum, develops in between markers D12S366 and D12S310) that was utero and presents shortly after birth as failure to pass common to all affected individuals. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Targeting the Tryptophan Hydroxylase 2 Gene for Functional Analysis in Mice and Serotonergic Differentiation of Embryonic Stem Cells
TARGETING THE TRYPTOPHAN HYDROXYLASE 2 GENE FOR FUNCTIONAL ANALYSIS IN MICE AND SEROTONERGIC DIFFERENTIATION OF EMBRYONIC STEM CELLS Inaugural-Dissertation to obtain the academic degree Doctor rerum naturalium (Dr. rer. nat.) submitted to the Department of Biology, Chemistry and Pharmacy of Freie Universität Berlin by Dana Kikic, M.Sc. in Molecular biology and Physiology from Nis June, 2009 The doctorate studies were performed in the research group of Prof. Michael Bader Molecular Biology of Peptide Hormones at Max-Delbrück-Center for Molecular Medicine in Berlin, Buch Mai 2005 - September 2008. 1st Reviewer: Prof. Michael Bader 2nd Reviewer: Prof. Udo Heinemann date of defence: 13. August 2009 ACKNOWLEDGMENTS Herewith, I would like to acknowledge the persons who made this thesis possible and without whom my initiation in the world of basic science research would not have the spin it has now, neither would my scientific illiteracy get the chance to eradicate. I am expressing my very personal gratitude and recognition to: Prof. Michael Bader, for an inexhaustible guidance in all the matters arising during the course of scientific work, for an instinct in defining and following the intellectual challenge and for letting me following my own, for necessary financial support, for defining the borders of reasonable and unreasonable, for an invaluable time and patience, and an amazing efficiency in supporting, motivating, reading, correcting and shaping my scientific language during the last four years. Prof. Harald Saumweber and Prof. Udo Heinemann, for taking over the academic supervision of the thesis, and for breathing in it a life outside the laboratory walls and their personal signature. -

Cyclin K Interacts with Β-Catenin to Induce Cyclin D1 Expression And
Theranostics 2020, Vol. 10, Issue 24 11144 Ivyspring International Publisher Theranostics 2020; 10(24): 11144-11158. doi: 10.7150/thno.42578 Research Paper Cyclin K interacts with β-catenin to induce Cyclin D1 expression and facilitates tumorigenesis and radioresistance in lung cancer Guojun Yao*, Jing Tang*, Xijie Yang, Ye Zhao, Rui Zhou, Rui Meng, Sheng Zhang, Xiaorong Dong, Tao Zhang, Kunyu Yang, Gang Wu and Shuangbing Xu Cancer Center, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China. *These authors contributed equally to this work. Corresponding author: Shuangbing Xu or Gang Wu, Cancer Center, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430022, China. E-mail: [email protected] or [email protected]. © The author(s). This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). See http://ivyspring.com/terms for full terms and conditions. Received: 2019.11.29; Accepted: 2020.08.24; Published: 2020.09.11 Abstract Rationale: Radioresistance remains the major cause of local relapse and distant metastasis in lung cancer. However, the underlying molecular mechanisms remain poorly defined. This study aimed to investigate the role and regulatory mechanism of Cyclin K in lung cancer radioresistance. Methods: Expression levels of Cyclin K were measured by immunohistochemistry in human lung cancer tissues and adjacent normal lung tissues. Cell growth and proliferation, neutral comet and foci formation assays, G2/M checkpoint and a xenograft mouse model were used for functional analyses. Gene expression was examined by RNA sequencing and quantitative real-time PCR. -

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -

Download Download
Supplementary Figure S1. Results of flow cytometry analysis, performed to estimate CD34 positivity, after immunomagnetic separation in two different experiments. As monoclonal antibody for labeling the sample, the fluorescein isothiocyanate (FITC)- conjugated mouse anti-human CD34 MoAb (Mylteni) was used. Briefly, cell samples were incubated in the presence of the indicated MoAbs, at the proper dilution, in PBS containing 5% FCS and 1% Fc receptor (FcR) blocking reagent (Miltenyi) for 30 min at 4 C. Cells were then washed twice, resuspended with PBS and analyzed by a Coulter Epics XL (Coulter Electronics Inc., Hialeah, FL, USA) flow cytometer. only use Non-commercial 1 Supplementary Table S1. Complete list of the datasets used in this study and their sources. GEO Total samples Geo selected GEO accession of used Platform Reference series in series samples samples GSM142565 GSM142566 GSM142567 GSM142568 GSE6146 HG-U133A 14 8 - GSM142569 GSM142571 GSM142572 GSM142574 GSM51391 GSM51392 GSE2666 HG-U133A 36 4 1 GSM51393 GSM51394 only GSM321583 GSE12803 HG-U133A 20 3 GSM321584 2 GSM321585 use Promyelocytes_1 Promyelocytes_2 Promyelocytes_3 Promyelocytes_4 HG-U133A 8 8 3 GSE64282 Promyelocytes_5 Promyelocytes_6 Promyelocytes_7 Promyelocytes_8 Non-commercial 2 Supplementary Table S2. Chromosomal regions up-regulated in CD34+ samples as identified by the LAP procedure with the two-class statistics coded in the PREDA R package and an FDR threshold of 0.5. Functional enrichment analysis has been performed using DAVID (http://david.abcc.ncifcrf.gov/)