A Comprehensive Analysis of the Expression of Crystallins in Mouse Retina Jinghua Xi Washington University School of Medicine in St
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Role of Autophagy in Eye Diseases
life Review The Role of Autophagy in Eye Diseases José A. Fernández-Albarral 1 , Esther de Julián-López 1, Carmen Soler-Domínguez 1, Rosa de Hoz 1,2,3 , Inés López-Cuenca 1 , Elena Salobrar-García 1,2,3 , José M. Ramírez 1,2,4 , María D. Pinazo-Durán 2,5, Juan J. Salazar 1,2,3,* and Ana I. Ramírez 1,2,3,* 1 Instituto de Investigaciones Oftalmológicas Ramón Castroviejo, Universidad Complutense de Madrid (UCM), 28040 Madrid, Spain; [email protected] (J.A.F.-A.); [email protected] (E.d.J.-L.); [email protected] (C.S.-D.); [email protected] (R.d.H.); [email protected] (I.L.-C.); [email protected] (E.S.-G.); [email protected] (J.M.R.) 2 OFTARED: Spanish net of Ophthalmic Pathology, Institute of Health Carlos III, 28029 Madrid, Spain; [email protected] 3 Departamento de Inmunología, Facultad de Óptica y Optometría, Oftalmología y ORL, UCM, 28037 Madrid, Spain 4 Departamento de Inmunología, Facultad de Medicina, Oftalmología y ORL. UCM, 28040 Madrid, Spain 5 Ophthalmic Research Unit “Santiago Grisolía”-FISABIO and Cellular and Molecular Ophthalmobiology Unit, University of Valencia, 46017 Valencia, Spain * Correspondence: [email protected] (J.J.S.); [email protected] (A.I.R.) Abstract: Autophagy is a catabolic process that ensures homeostasis in the cells of our organism. It plays a crucial role in protecting eye cells against oxidative damage and external stress factors. Ocular pathologies of high incidence, such as age-related macular degeneration, cataracts, glaucoma, and diabetic retinopathy are of multifactorial origin and are associated with genetic, environmental factors, age, and oxidative stress, among others; the latter factor is one of the most influential in ocular Citation: Fernández-Albarral, J.A.; diseases, directly affecting the processes of autophagy activity. -

Identification of the Binding Partners for Hspb2 and Cryab Reveals
Brigham Young University BYU ScholarsArchive Theses and Dissertations 2013-12-12 Identification of the Binding arP tners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non- Redundant Roles for Small Heat Shock Proteins Kelsey Murphey Langston Brigham Young University - Provo Follow this and additional works at: https://scholarsarchive.byu.edu/etd Part of the Microbiology Commons BYU ScholarsArchive Citation Langston, Kelsey Murphey, "Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins" (2013). Theses and Dissertations. 3822. https://scholarsarchive.byu.edu/etd/3822 This Thesis is brought to you for free and open access by BYU ScholarsArchive. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston A thesis submitted to the faculty of Brigham Young University in partial fulfillment of the requirements for the degree of Master of Science Julianne H. Grose, Chair William R. McCleary Brian Poole Department of Microbiology and Molecular Biology Brigham Young University December 2013 Copyright © 2013 Kelsey Langston All Rights Reserved ABSTRACT Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactors and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston Department of Microbiology and Molecular Biology, BYU Master of Science Small Heat Shock Proteins (sHSP) are molecular chaperones that play protective roles in cell survival and have been shown to possess chaperone activity. -
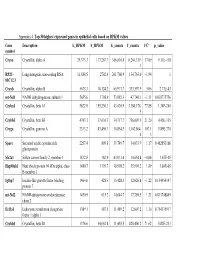
Appendix 4. Top 50 Highest Expressed Genes in Epithelial Cells Based on RPKM Values
Appendix 4. Top 50 highest expressed genes in epithelial cells based on RPKM values Gene Description E_RPKM F_RPKM E_counts F_counts FC* p_value symbol Cryaa Crystallin, alpha A 29,373.3 177,267.7 366,616.4 6,264,319. 17.09 9.11E-118 1 RP23– Long intergenic non-coding RNA 11,888.5 2702.4 261,760.9 134,763.0 −1.94 1 81C12.3 Cryab Crystallin, alpha B 5673.3 10,124.2 65,971.7 333,597.9 5.06 2.71E-43 mt-Nd1 NADH dehydrogenase, subunit 1 5655.6 1798.9 53,082.3 47,748.1 −1.11 0.838775756 Cryba1 Crystallin, beta A1 5622.0 155,230.3 43,420.9 3,380,176. 77.85 1.34E-240 5 Crybb3 Crystallin, beta B3 4743.1 37,636.3 34,717.7 736,007.9 21.20 4.45E-135 Cryga Crystallin, gamma A 2333.2 83,496.3 10,854.5 1,162,864. 107.1 5.89E-270 6 3 Sparc Secreted acidic cysteine rich 2257.4 809.8 39,749.7 34,033.9 −1.17 0.462853166 glycoprotein Slc2a1 Solute carrier family 2, member 1 1832.8 162.9 43,031.4 10,654.8 −4.04 1.67E-05 Hsp90ab1 Heat shock protein 90 kDa alpha, class 1480.7 1139.7 18,998.2 35,901.2 1.89 3.84E-05 B member 1 Igfbp7 Insulin-like growth factor binding 1464.6 428.3 15,428.3 12,626.8 −1.22 0.154954147 protein 7 mt-Nd2 NADH-ubiquinone oxidoreductase 1450.9 615.2 14,644.7 17,789.5 1.21 0.833748849 chain 2 Eef1a1 Eukaryotic translation elongation 1389.1 587.5 11,489.2 12,607.2 1.10 0.754135917 factor 1 alpha 1 Crybb1 Crystallin, beta B1 1376.6 34,662.8 11,455.5 820,406.2 71.62 5.82E-233 Htra3 HtrA serine peptidase 3 1338.6 162.0 23,197.6 6433.9 −3.61 3.93E-05 Gnb2l1 Guanine nucleotide-binding protein 1293.3 670.1 14,495.1 21,652.1 1.49 0.001685952 -

CRYGB Rabbit Pab
Leader in Biomolecular Solutions for Life Science CRYGB Rabbit pAb Catalog No.: A14569 Basic Information Background Catalog No. Crystallins are separated into two classes: taxon-specific, or enzyme, and ubiquitous. A14569 The latter class constitutes the major proteins of vertebrate eye lens and maintains the transparency and refractive index of the lens. Since lens central fiber cells lose their Observed MW nuclei during development, these crystallins are made and then retained throughout 21kDa life, making them extremely stable proteins. Mammalian lens crystallins are divided into alpha, beta, and gamma families; beta and gamma crystallins are also considered as a Calculated MW superfamily. Alpha and beta families are further divided into acidic and basic groups. 20kDa Seven protein regions exist in crystallins: four homologous motifs, a connecting peptide, and N- and C-terminal extensions. Gamma-crystallins are a homogeneous group of Category highly symmetrical, monomeric proteins typically lacking connecting peptides and terminal extensions. They are differentially regulated after early development. Four Primary antibody gamma-crystallin genes (gamma-A through gamma-D) and three pseudogenes (gamma-E, gamma-F, gamma-G) are tandemly organized in a genomic segment as a Applications gene cluster. Whether due to aging or mutations in specific genes, gamma-crystallins WB have been involved in cataract formation. Cross-Reactivity Mouse, Rat Recommended Dilutions Immunogen Information WB 1:500 - 1:2000 Gene ID Swiss Prot 1419 P07316 Immunogen Recombinant fusion protein containing a sequence corresponding to amino acids 80-135 of human CRYGB (NP_005201.2). Synonyms CRYGB;CRYG2;CTRCT39 Contact Product Information www.abclonal.com Source Isotype Purification Rabbit IgG Affinity purification Storage Store at -20℃. -

Supp Table 1.Pdf
Upregulated genes in Hdac8 null cranial neural crest cells fold change Gene Symbol Gene Title 134.39 Stmn4 stathmin-like 4 46.05 Lhx1 LIM homeobox protein 1 31.45 Lect2 leukocyte cell-derived chemotaxin 2 31.09 Zfp108 zinc finger protein 108 27.74 0710007G10Rik RIKEN cDNA 0710007G10 gene 26.31 1700019O17Rik RIKEN cDNA 1700019O17 gene 25.72 Cyb561 Cytochrome b-561 25.35 Tsc22d1 TSC22 domain family, member 1 25.27 4921513I08Rik RIKEN cDNA 4921513I08 gene 24.58 Ofa oncofetal antigen 24.47 B230112I24Rik RIKEN cDNA B230112I24 gene 23.86 Uty ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome 22.84 D8Ertd268e DNA segment, Chr 8, ERATO Doi 268, expressed 19.78 Dag1 Dystroglycan 1 19.74 Pkn1 protein kinase N1 18.64 Cts8 cathepsin 8 18.23 1500012D20Rik RIKEN cDNA 1500012D20 gene 18.09 Slc43a2 solute carrier family 43, member 2 17.17 Pcm1 Pericentriolar material 1 17.17 Prg2 proteoglycan 2, bone marrow 17.11 LOC671579 hypothetical protein LOC671579 17.11 Slco1a5 solute carrier organic anion transporter family, member 1a5 17.02 Fbxl7 F-box and leucine-rich repeat protein 7 17.02 Kcns2 K+ voltage-gated channel, subfamily S, 2 16.93 AW493845 Expressed sequence AW493845 16.12 1600014K23Rik RIKEN cDNA 1600014K23 gene 15.71 Cst8 cystatin 8 (cystatin-related epididymal spermatogenic) 15.68 4922502D21Rik RIKEN cDNA 4922502D21 gene 15.32 2810011L19Rik RIKEN cDNA 2810011L19 gene 15.08 Btbd9 BTB (POZ) domain containing 9 14.77 Hoxa11os homeo box A11, opposite strand transcript 14.74 Obp1a odorant binding protein Ia 14.72 ORF28 open reading -

Related Macular Degeneration and Cutis Laxa
UvA-DARE (Digital Academic Repository) Genetic studies of age-related macular degeneration Baas, D.C. Publication date 2012 Document Version Final published version Link to publication Citation for published version (APA): Baas, D. C. (2012). Genetic studies of age-related macular degeneration. General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:05 Oct 2021 G������ S������ �� A��-������� M������ D����������� D����������� M������ G������ S������ �� A��-������� | 2012 D�������� C. B��� G������ S������ �� A��-������� M������ D����������� D�������� C. B��� cover.indd 1 31-10-12 08:36 Genetic Studies of Age-related Macular Degeneration Dominique C. Baas Chapter 0.indd 1 23-10-12 19:24 The research described in this thesis was conducted at the Netherlands Institute for Neuroscience (NIN), an institute of the Royal Netherlands Academy of Arts and Sciences, Department of Clinical and Molecular Ophthalmogenetics, Amsterdam, The Netherlands. -

Method for Diagnosing Non-Small Cell Lung Cancers
(19) TZZ Z _ T (11) EP 2 270 221 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 05.01.2011 Bulletin 2011/01 C12Q 1/68 (2006.01) C12N 15/12 (2006.01) G01N 33/50 (2006.01) C12N 15/11 (2006.01) (2006.01) (2006.01) (21) Application number: 10010329.0 A61P 35/00 A61K 39/00 (22) Date of filing: 22.09.2003 (84) Designated Contracting States: (72) Inventors: AT BE BG CH CY CZ DE DK EE ES FI FR GB GR • Nakamura, Yusuke HU IE IT LI LU MC NL PT RO SE SI SK TR Yokohama-shi Kanagawa 225-0011 (JP) (30) Priority: 30.09.2002 US 414673 P • Daigo, Yataro 28.02.2003 US 451374 P Yokohama-shi 28.04.2003 US 466100 P Kanagawa 222-0031 (JP) • Nakatsuru, Shuichi (62) Document number(s) of the earlier application(s) in Saitama-shi accordance with Art. 76 EPC: Saitama 338-0002 (JP) 06022167.8 / 1 743 947 03753941.8 / 1 551 998 (74) Representative: Vossius & Partner Siebertstrasse 4 (71) Applicant: Oncotherapy Science, Inc. 81675 München (DE) Kawasaki-shi Kanagawa 213-0012 (JP) Remarks: This application was filed on 23-09-2010 as a divisional application to the application mentioned under INID code 62. (54) Method for diagnosing non-small cell lung cancers (57) Disclosed are methods for detecting non-small cancerous tissues are provided. Also disclosed are meth- cell lung cancer using differentially expressed genes. ods of identifying compounds for treating and preventing Furthermore, novel human genes whose expression is non-small cell lung cancer. -

Congenital Cataracts Due to a Novel 2‑Bp Deletion in CRYBA1/A3
1614 MOLECULAR MEDICINE REPORTS 10: 1614-1618, 2014 Congenital cataracts due to a novel 2‑bp deletion in CRYBA1/A3 JING ZHANG1, YANHUA ZHANG1, FANG FANG1, WEIHONG MU1, NING ZHANG2, TONGSHUN XU3 and QINYING CAO1 1Prenatal Diagnosis Center, Shijiazhuang Obstetrics and Gynecology Hospital; 2Department of Cardiology, The Second Hospital of Hebei Medical University; 3Department of Surgery, Shijiazhuang Obstetrics and Gynecology Hospital, Shijiazhuang, Hebei, P.R. China Received September 22, 2013; Accepted April 11, 2014 DOI: 10.3892/mmr.2014.2324 Abstract. Congenital cataracts, which are a clinically and located in the eye lens. The major human crystallins comprise genetically heterogeneous group of eye disorders, lead to 90% of protein in the mature lens and contain two different visual impairment and are a significant cause of blindness superfamilies: the small heat‑shock proteins (α-crystallins) in childhood. A major proportion of the causative mutations and the βγ-crystallins. for congenital cataracts are found in crystallin genes. In the In this study a functional candidate approach was used present study, a novel deletion mutation (c.590-591delAG) in to investigate the known crystallin genes, including CRYAA, exon 6 of CRYBA1/A3 was identified in a large family with CRYAB, CRYBA1/A3, CRYBB1, CRYBB2, CRYGC, CRYGD autosomal dominant congenital cataracts. An increase in and CRYGS, in which a major proportion of the mutations local hydrophobicity was predicted around the mutation site; identified in a large family with congenital cataracts were however, further studies are required to determine the exact found. effect of the mutation on βA1/A3-crystallin structure and function. To the best of our knowledge, this is the first report Subjects and methods of an association between a frameshift mutation in exon 6 of CRYBA1/A3 and congenital cataracts. -

Congenital Eye Disorders Gene Panel
Congenital eye disorders gene panel Contact details Introduction Regional Genetics Service Ocular conditions are highly heterogeneous and show considerable phenotypic overlap. 1 in Levels 4-6, Barclay House 2,500 children in the UK are diagnosed as blind or severely visually impaired by the time they 37 Queen Square reach one year old. As many as half of these cases are likely to be inherited and remain undiagnosed due to the vast number of genes involved in these conditions. Many congenital London, WC1N 3BH eye disorders causing visual impairment or blindness at birth or progressive visual impairment T +44 (0) 20 7762 6888 also include syndromic conditions involving additional metabolic, developmental, physical or F +44 (0) 20 7813 8578 sensory abnormalities. Gene panels offer the enhanced probability of diagnosis as a very large number of genes can be interrogated. Samples required Ocular birth defects include all inheritance modalities. Autosomal dominant and recessive 5ml venous blood in plastic EDTA diseases as well as X-linked dominant and recessive diseases are seen. These conditions can bottles (>1ml from neonates) also be caused by de novo variants. Prenatal testing must be arranged Referrals in advance, through a Clinical Genetics department if possible. Patients presenting with a phenotype appropriate for the requested sub-panel Amniotic fluid or CV samples Referrals will be accepted from clinical geneticists and consultants in ophthalmology. should be sent to Cytogenetics for Prenatal testing dissecting and culturing, with instructions to forward the sample Prenatal diagnosis may be offered as appropriate where pathogenic variants have been to the Regional Molecular Genetics identified in accordance with expected inheritance pattern and where appropriate parental laboratory for analysis testing and counselling has been conducted. -

Γd-Crystallin Aggregation Precursor 1 an Internal Disulfide Locks A
γD-crystallin aggregation precursor An internal disulfide locks a misfolded aggregation-prone intermediate in cataract-linked mutants of human γD-crystallin Eugene Serebryany*1, Jaie C. Woodard*2, Bharat V. Adkar2, Mohammed Shabab1, Jonathan A. King†1, and Eugene I. Shakhnovich†2 1Department of Biology, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139 2Department of Chemistry and Chemical Biology, Harvard University, Cambridge, Massachusetts 02138 *Equal contribution. †To whom correspondence should be addressed: [email protected]; [email protected] Running title: γD-crystallin aggregation precursor Considerable mechanistic insight has been gained Partially unfolded or misfolded, aggregation-prone into amyloid aggregation; however, a large class of protein conformational states are linked to a wide non-amyloid protein aggregates are considered array of age-related protein misfolding diseases. The “amorphous,” and in most cases little is known best studied of these conditions include amyotrophic about their mechanisms. Amorphous aggregation of lateral sclerosis (superoxide dismutase), Parkinson’s γ-crystallins in the eye lens causes a widespread disease (α-synuclein), serpinopathies (α1-antitrypsin), disease of aging, cataract. We combined simulations cancers with P53 and P21 tumor suppressor defects, and experiments to study the mechanism of and lens cataract (crystallins) (1-5). Some of these aggregation of two γD-crystallin mutants, W42R and aggregates contain the well-known amyloid W42Q – the former a congenital cataract mutation, structure, and others do not; even in cases where and the latter a mimic of age-related oxidative amyloid is the final aggregated state, oligomers, damage. We found that formation of an internal prefibrillar species, and amorphous aggregates are disulfide was necessary and sufficient for often closely linked to pathology (6,7). -

Transgenic Zebrafish Models Reveal Distinct Molecular Mechanisms for Cataract-Linked Αa-Crystallin Mutants
bioRxiv preprint doi: https://doi.org/10.1101/364125; this version posted July 8, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Transgenic Zebrafish Models Reveal Distinct Molecular Mechanisms for Cataract-linked αA-Crystallin Mutants Shu-Yu Wu, Ping Zou, Sanjay Mishra, Hassane S Mchaourab* Department of Molecular Physiology and Biophysics, Vanderbilt University, Nashville, TN 37232, USA Running title: Distinct mechanisms of α-crystallin mutations * Corresponding author: Hassane Mchaourab 741 Light Hall 2215 Garland Avenue Molecular Physiology & Biophysics Nashville, TN 37232 Office: 615.322.3307 Fax: 615.322.7236 Email: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/364125; this version posted July 8, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Mutations in the small heat shock proteins a-crystallins have been linked to autosomal dominant cataracts in humans. Extensive studies in vitro have revealed a spectrum of alterations to the structure and function of these proteins including shifts in the size of the oligomer, modulation of subunit exchange and modification of their affinity to client proteins. Although mouse models of these mutants were instrumental in identifying changes in cellular proliferation and lens development, a direct comparative analysis of their effects on lens proteostasis has not been performed. -

Supplementary Materials
Supplementary materials Gene Nucleotide Amino acid change Clinical phenotype Ref symbol change CRYAA c.35G>T p. R12L lens protein gene [1] CRYAB c.32G>A p. R11H lens protein gene [2] CRYBA1 c.279-281delG p.ΔG91 lens protein gene [3] AG CRYBA4 c.206T>C p. L69P lens protein gene [4] CRYBB1 c.658G>T p. G220X lens protein gene [5] CRYBB2 c.563G>A p. R188H lens protein gene [6] CRYBB3 c.314G>A p. R105Q lens protein gene [7] CRYGA c.196T>C p. Y66H lens protein gene [8] CRYGB c.449G>T p. G150V lens protein gene [8] CRYGC c.385G>T p. G129C lens protein gene [9] CRYGD c.70C>A p. P24T lens protein gene [10] CRYGS c.53G>T :p.G18V lens protein gene [11] GJA3 c.188A>G p.N63S membrane protein [12] gene GJA8 c.262C>T p.P88S membrane protein [13] gene BFSP1 c736-1384_c.9 T246fsX7 cytoskeleton protein [14] 57-66del gene BFSP2 c.1091G>A p.G364D cytoskeleton protein [15] gene PAX6 c.307C>T p.R103X developmental [16] regulatory protein gen PITX3 c.38G>A p.S13N developmental [17] regulatory protein gen HSF4 c.524G>C p.R175P developmental [18] regulatory protein gen MAF c.863G>C p.R288P developmental [19] regulatory protein gen CHMP4B c.481G>A p. E161K chromatin modified [20] protein gene EPHA2 c.2842G>T p. G948W tyrosine kinase [21] receptor gene COL4A1 c.2345G>C p. G782A syndrome-related [22] genes FTL c.160G>A p.E54K developmental [23] regulatory protein gen GALK1 c.416T>C p.