Omics in Myopia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
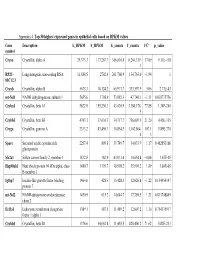
Appendix 4. Top 50 Highest Expressed Genes in Epithelial Cells Based on RPKM Values
Appendix 4. Top 50 highest expressed genes in epithelial cells based on RPKM values Gene Description E_RPKM F_RPKM E_counts F_counts FC* p_value symbol Cryaa Crystallin, alpha A 29,373.3 177,267.7 366,616.4 6,264,319. 17.09 9.11E-118 1 RP23– Long intergenic non-coding RNA 11,888.5 2702.4 261,760.9 134,763.0 −1.94 1 81C12.3 Cryab Crystallin, alpha B 5673.3 10,124.2 65,971.7 333,597.9 5.06 2.71E-43 mt-Nd1 NADH dehydrogenase, subunit 1 5655.6 1798.9 53,082.3 47,748.1 −1.11 0.838775756 Cryba1 Crystallin, beta A1 5622.0 155,230.3 43,420.9 3,380,176. 77.85 1.34E-240 5 Crybb3 Crystallin, beta B3 4743.1 37,636.3 34,717.7 736,007.9 21.20 4.45E-135 Cryga Crystallin, gamma A 2333.2 83,496.3 10,854.5 1,162,864. 107.1 5.89E-270 6 3 Sparc Secreted acidic cysteine rich 2257.4 809.8 39,749.7 34,033.9 −1.17 0.462853166 glycoprotein Slc2a1 Solute carrier family 2, member 1 1832.8 162.9 43,031.4 10,654.8 −4.04 1.67E-05 Hsp90ab1 Heat shock protein 90 kDa alpha, class 1480.7 1139.7 18,998.2 35,901.2 1.89 3.84E-05 B member 1 Igfbp7 Insulin-like growth factor binding 1464.6 428.3 15,428.3 12,626.8 −1.22 0.154954147 protein 7 mt-Nd2 NADH-ubiquinone oxidoreductase 1450.9 615.2 14,644.7 17,789.5 1.21 0.833748849 chain 2 Eef1a1 Eukaryotic translation elongation 1389.1 587.5 11,489.2 12,607.2 1.10 0.754135917 factor 1 alpha 1 Crybb1 Crystallin, beta B1 1376.6 34,662.8 11,455.5 820,406.2 71.62 5.82E-233 Htra3 HtrA serine peptidase 3 1338.6 162.0 23,197.6 6433.9 −3.61 3.93E-05 Gnb2l1 Guanine nucleotide-binding protein 1293.3 670.1 14,495.1 21,652.1 1.49 0.001685952 -

Supp Table 1.Pdf
Upregulated genes in Hdac8 null cranial neural crest cells fold change Gene Symbol Gene Title 134.39 Stmn4 stathmin-like 4 46.05 Lhx1 LIM homeobox protein 1 31.45 Lect2 leukocyte cell-derived chemotaxin 2 31.09 Zfp108 zinc finger protein 108 27.74 0710007G10Rik RIKEN cDNA 0710007G10 gene 26.31 1700019O17Rik RIKEN cDNA 1700019O17 gene 25.72 Cyb561 Cytochrome b-561 25.35 Tsc22d1 TSC22 domain family, member 1 25.27 4921513I08Rik RIKEN cDNA 4921513I08 gene 24.58 Ofa oncofetal antigen 24.47 B230112I24Rik RIKEN cDNA B230112I24 gene 23.86 Uty ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome 22.84 D8Ertd268e DNA segment, Chr 8, ERATO Doi 268, expressed 19.78 Dag1 Dystroglycan 1 19.74 Pkn1 protein kinase N1 18.64 Cts8 cathepsin 8 18.23 1500012D20Rik RIKEN cDNA 1500012D20 gene 18.09 Slc43a2 solute carrier family 43, member 2 17.17 Pcm1 Pericentriolar material 1 17.17 Prg2 proteoglycan 2, bone marrow 17.11 LOC671579 hypothetical protein LOC671579 17.11 Slco1a5 solute carrier organic anion transporter family, member 1a5 17.02 Fbxl7 F-box and leucine-rich repeat protein 7 17.02 Kcns2 K+ voltage-gated channel, subfamily S, 2 16.93 AW493845 Expressed sequence AW493845 16.12 1600014K23Rik RIKEN cDNA 1600014K23 gene 15.71 Cst8 cystatin 8 (cystatin-related epididymal spermatogenic) 15.68 4922502D21Rik RIKEN cDNA 4922502D21 gene 15.32 2810011L19Rik RIKEN cDNA 2810011L19 gene 15.08 Btbd9 BTB (POZ) domain containing 9 14.77 Hoxa11os homeo box A11, opposite strand transcript 14.74 Obp1a odorant binding protein Ia 14.72 ORF28 open reading -

A Comprehensive Analysis of the Expression of Crystallins in Mouse Retina Jinghua Xi Washington University School of Medicine in St
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2003 A comprehensive analysis of the expression of crystallins in mouse retina Jinghua Xi Washington University School of Medicine in St. Louis Rafal Farjo University of Michigan - Ann Arbor Shigeo Yoshida University of Michigan - Ann Arbor Timothy S. Kern Case Western Reserve University Anand Swaroop University of Michigan - Ann Arbor See next page for additional authors Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs Recommended Citation Xi, Jinghua; Farjo, Rafal; Yoshida, Shigeo; Kern, Timothy S.; Swaroop, Anand; and Andley, Usha P., ,"A comprehensive analysis of the expression of crystallins in mouse retina." Molecular Vision.9,. 410-419. (2003). https://digitalcommons.wustl.edu/open_access_pubs/1801 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. Authors Jinghua Xi, Rafal Farjo, Shigeo Yoshida, Timothy S. Kern, Anand Swaroop, and Usha P. Andley This open access publication is available at Digital Commons@Becker: https://digitalcommons.wustl.edu/open_access_pubs/1801 Molecular Vision 2003; 9:410-9 <http://www.molvis.org/molvis/v9/a53> © 2003 Molecular Vision Received 28 May 2003 | Accepted 19 August 2003 | Published 28 August 2003 A comprehensive analysis of the expression of crystallins in mouse retina Jinghua Xi,1 Rafal Farjo,3 Shigeo Yoshida,3 Timothy S. Kern,5 Anand Swaroop,3,4 Usha P. Andley1,2 Departments of 1Ophthalmology and Visual Sciences and 2Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. -

Oxidative Stress and Inflammation in Hepatic Diseases
Review Oxidative Stress and Inflammation in Hepatic Diseases: Therapeutic Possibilities of N-Acetylcysteine Kívia Queiroz de Andrade 1, Fabiana Andréa Moura 1,2, John Marques dos Santos 3, Orlando Roberto Pimentel de Araújo 3, Juliana Célia de Farias Santos 2 and Marília Oliveira Fonseca Goulart 3,* Received: 10 October 2015; Accepted: 4 December 2015; Published: 18 December 2015 Academic Editor: Guido Haenen 1 Pós Graduação em Ciências da Saúde (PPGCS), Campus A. C. Simões, Tabuleiro dos Martins, 57072-970 Maceió, AL, Brazil; [email protected] (K.Q.A.); [email protected] (F.A.M.) 2 Faculdade de Nutrição/Universidade Federal de Alagoas (FANUT/UFAL), Campus A. C. Simões, Tabuleiro dos Martins, 57072-970 Maceió, AL, Brazil; [email protected] 3 Instituto de Química e Biotecnologia (IQB), Universidade Federal de Alagoas (UFAL), Campus A. C. Simões, Tabuleiro dos Martins, 57072-970 Maceió, AL, Brazil; [email protected] (J.M.S.); [email protected] (O.R.P.A.) * Correspondence: [email protected]; Tel.: +55-82-98818-0463 Abstract: Liver disease is highly prevalent in the world. Oxidative stress (OS) and inflammation are the most important pathogenetic events in liver diseases, regardless the different etiology and natural course. N-acetyl-L-cysteine (the active form) (NAC) is being studied in diseases characterized by increased OS or decreased glutathione (GSH) level. NAC acts mainly on the supply of cysteine for GSH synthesis. The objective of this review is to examine experimental and clinical studies that evaluate the antioxidant and anti-inflammatory roles of NAC in attenuating markers of inflammation and OS in hepatic damage. -

Identification of a Distinct Metabolomic Subtype of Sporadic ALS Patients
bioRxiv preprint doi: https://doi.org/10.1101/416396; this version posted September 13, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Identification of a Distinct Metabolomic Subtype of Sporadic ALS Patients Running title – Increased cysteine and glucose metabolism in sALS cases Qiuying Chen, PhD1*, Davinder Sandhu, M.S.1*, Csaba Konrad, PhD2, Dipa Roychoudhury, PhD3, Benjamin I. Schwartz1, Roger R. Cheng1, Kirsten Bredvik2, Hibiki Kawamata, PhD2, Elizabeth L. Calder, PhD4, Lorenz Studer, MD4, Steven. M. Fischer3, Giovanni Manfredi MD/PhD2* and Steven. S. Gross, PhD1* *Joint first/senior authors 1Department of Pharmacology, Weill Cornell Medicine, New York, NY, USA 2Brain and Mind Research Institute, Weill Cornell Medicine, New York, NY, USA 3Agilent Technology, Santa Clara, CA, USA 4The Center for Stem Cell Biology, Sloan-Kettering Institute for Cancer Center, New York, NY. Keywords: sporadic amyotrophic lateral sclerosis, metabolomics, stable isotope tracing, trans-sulfuration, hypermetabolism, disease stratification, energy metabolism Co-Corresponding Authors: Steven S. Gross, PhD Department of Pharmacology Weill Cornell Medicine Phone: 212-746-6257 Email: [email protected] Giovanni Manfredi, MD/PhD Brain and Mind Research Institute Weill Cornell Medicine Phone: 646-962-8172 Email: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/416396; this version posted September 13, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Number of characters: Title --- 94 Running head---48 Number of words: Title --- 13 Running head --- 8 Abstract---232 Introduction --- 549 Discussion --- 1126 Body of manuscript --- 5706 Number of figures: 8 Number of color figures: 8 Number of tables:1 2 bioRxiv preprint doi: https://doi.org/10.1101/416396; this version posted September 13, 2018. -

Supplemental Table 1. Differences in Fasting Metabolites (All) by CKD Status in Plasma
Supplemental table 1. Differences in fasting metabolites (all) by CKD status in plasma. Metabolites in bold show hits controlling FDR at 10%, in a model that controls for age, sex, race (white versus not), and weight. Ordered by subclasses of metabolites followed by adjusted percent difference between CKD and controls and adjusted p-value for difference. Metabolite % Difference p-value q-value Pathway in CKD, compared with controls Cystine (241.1 / 120.0) 33 2.60E-09 1.52E-08 Amino Acid Proline (116.1 / 70.0) 20 0.000195 0.000692 Amino Acid 2-Hydroxyisovaleric Acid (117.0 / 71.0) 47 0.000331 0.001139 Amino Acid Tyrosine (182.1 / 136.0) -13 0.000633 0.002014 Amino Acid Serine (106.0 / 60.0) -8 0.029507 0.065132 Amino Acid Glycine (76.0 / 30.1) 13 0.032003 0.06842 Amino Acid Leucine (132.1 / 86.0) -5 0.067499 0.139499 Amino Acid 5-Aminovaleric Acid (118.0 / 55.0) -4 0.127339 0.253324 Amino Acid Threonine (120.1 / 74.0 (2)) -7 0.145125 0.281179 Amino Acid Glutamine (147.1 / 84.0) 3 0.169291 0.318061 Amino Acid Valine (118.1 / 72.0) -4 0.201091 0.36138 Amino Acid Arginine (175.1 / 70.0) 5 0.209045 0.362752 Amino Acid Sarcosine (89.9 / 44.0) -7 0.209395 0.362752 Amino Acid Cadaverine (103.0 / 86.0) 4 0.281896 0.459935 Amino Acid Histidine (156.1 / 110.0) 3 0.319987 0.495386 Amino Acid Alanine (90.0 / 44.0) 5 0.330111 0.499193 Amino Acid iso-Leucine (132.1 / 86.0 (2)) -3 0.336431 0.50262 Amino Acid Phenylalanine (166.1 / 120.0) 2 0.461445 0.61526 Amino Acid Methionine (150.1 / 61.0) -3 0.516557 0.681415 Amino Acid Pipecolate (130.0 / 84.0) -2 -

Taurine Supplementation Improves the Utilization of Sulfur
Available online at www.sciencedirect.com Journal of Nutritional Biochemistry 20 (2009) 132–139 Taurine supplementation improves the utilization of sulfur-containing amino acids in rats continually administrated alcohol Hui-Ting Yanga, Yi-Wen Chienb, Jen-Horng Tsenc, Ching-Chien Changb,d, ⁎ ⁎ Jer-Hwa Change, ,1, Shih-Yi Huangb, ,1 aSchool of Pharmaceutical Science, Taipei Medical University, Taipei bSchool of Nutrition and Health Sciences, Taipei Medical University, Taipei cSchool of Nutrition, China Medical University, Taichung, Taiwan dGeneral Education Center, Northern Taiwan Institute of Science and Technology, Taipei eDepartment of Internal Medicine, Taipei Medical University Wan Fang Hospital, Taipei, Taiwan Received 27 September 2007; received in revised form 18 December 2007; accepted 4 January 2008 Abstract The main purpose of this study was to evaluate changes in brain sulfur-containing amino acid (SCAA) metabolism to determine whether taurine intervened under continuous alcohol intake. We fed 80 male Sprague–Dawley rats 30% alcohol-containing water for 4 weeks. Eighty animals were divided into two groups (with or without 2 g/kg body weight taurine supplementation), and five were killed every week in each group for monitoring SCAA changes in the brain, liver, kidneys and heart. Results indicated that the plasma alcohol concentration increased from Weeks 1–4; however, animals with taurine supplementation showed a lower plasma concentration of ethanol in Week 2. As to SCAA concentrations, cysteine and taurine were both lower after a week of alcohol ingestion in the brain and plasma; the same declining trend was shown in the liver in Week 2. In contrast, plasma and hepatic concentrations of homocysteine were elevated in Week 2, and the plasma S-adenosylmethionine (SAM)/S-adenosylhomocysteine (SAH) ratio also decreased in Week 1. -

Combined Metabolic Activators Improve Cognitive Functions Without Altering Motor
medRxiv preprint doi: https://doi.org/10.1101/2021.07.28.21261293; this version posted August 4, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . Combined Metabolic Activators Improve Cognitive Functions without Altering Motor Scores in Parkinson’s Disease Burak Yulug1,#, Ozlem Altay2,#, Xiangyu Li2, #, Lutfu Hanoglu3, Seyda Cankaya1, Simon Lam4, Hong Yang2, Ebru Coskun3, Ezgi İdil1, Rahim Nogaylar1, Ahmet Hacımuftuoglu5, Muhammad Arif2, Saeed Shoaie2,4, Cheng Zhang2.6, Jens Nielsen7, Hasan Turkez8, Jan Borén9, Mathias Uhlén2,*, Adil Mardinoglu2,4,* 1Department of Neurology and Neuroscience, Faculty of Medicine, Alanya Alaaddin Keykubat University, Antalya, Turkey 2Science for Life Laboratory, KTH - Royal Institute of Technology, Stockholm, Sweden 3Department of Neurology, Faculty of Medicine, Istanbul Medipol University, Istanbul, Turkey 4Centre for Host-Microbiome Interactions, Faculty of Dentistry, Oral & Craniofacial Sciences, King’s College London, London, United Kingdom 5Department of Medical Pharmacology, Faculty of Medicine, Atatürk University, Erzurum, Turkey. 6School of Pharmaceutical Sciences, Zhengzhou University, Zhengzhou, PR China 7Department of Biology and Biological Engineering, Chalmers University of Technology, Gothenburg, Sweden 8Department of Medical Biology, Faculty of Medicine, Atatürk University, Erzurum, Turkey 9Department -

The Biosynthesis Reaction of Hypotaurine to Taurine
UNIVERSITY OF CENTRAL OKLAHOMA Edmond, Oklahoma Jackson College of Graduate Studies The Biosynthesis Reaction of Hypotaurine to Taurine A THESIS SUBMITTED TO THE GRADUATE FACULTY In partial fulfillment of the requirements For the degree of MASTER OF SCIENCE IN BIOLOGY By Roxanna Q. Grove Edmond, Oklahoma 2018 Acknowledgments Working on this project has been a period of intense learning for me, not only in the scientific arena, but also on a personal level. Writing this thesis has had a significant impact on me. I would like to reflect on people who have been supported and helped me so much throughout this period. First of all, I would like to express my gratitude toward my advisor, Dr. Steven J. Karpowicz, for his devotion, inspiration, and guidance. I am so grateful to have the opportunity to work with such an intelligent, dedicated, and patient professor. I appreciate his vast knowledge and skills in many areas such as biochemistry, genetics, and bioinformatics, and his assistance in writing this thesis. I would like to thank the other members of my committee, Dr. Nikki Seagraves, Dr. Hari Kotturi, and Dr. Lilian Chooback, for their guidance, support, and for providing materials throughout this project. An exceptional thanks go to Dr. John Bowen of the Department of Chemistry for advice in the analytical laboratory and Dr. Susan L. Nimmo from the Department of Chemistry and Biochemistry at the University of Oklahoma for assistance with NMR. This project was supported by funding from the College of Mathematics and Science and a Research, Creative, and Scholarly Activities (RCSA) grant from the Office of High Impact Practices at UCO. -

Cysteamine: an Old Drug with New Potential
Drug Discovery Today Volume 18, Numbers 15/16 August 2013 REVIEWS POST SCREEN Cysteamine: an old drug with new potential Reviews 1,2 3 1,2,4 Martine Besouw , Rosalinde Masereeuw , Lambert van den Heuvel and 1,2 Elena Levtchenko 1 Department of Pediatric Nephrology, University Hospitals Leuven, Leuven, Belgium 2 Laboratory of Pediatrics, Department of Development & Regeneration, Catholic University Leuven, Belgium 3 Department of Pharmacology and Toxicology, Radboud University Nijmegen Medical Centre, Nijmegen Centre for Molecular Life Sciences, Nijmegen, The Netherlands 4 Laboratory for Genetic, Endocrine and Metabolic Disorders, Radboud University Nijmegen Medical Centre, Nijmegen, The Netherlands Cysteamine is an amino thiol with the chemical formula HSCH2CH2NH2. Endogenously, cysteamine is derived from coenzyme A degradation, although its plasma concentrations are low. Most experience with cysteamine as a drug originates from the field of the orphan disease cystinosis, in which cysteamine is prescribed to decrease intralysosomal cystine accumulation. However, over the years, the drug has been used for several other applications both in vitro and in vivo. In this article, we review the different applications of cysteamine, ending with an overview of ongoing clinical trials for new indications, such as neurodegenerative disorders and nonalcoholic fatty liver disease (NAFLD). The recent development of an enteric-coated cysteamine formulation makes cysteamine more patient friendly and will extend its applicability for both old and new indications. Endogenous cysteamine production cysteine, it is oxidized into taurine by hypotaurine dehydrogenase Cysteamine (synonyms: b-mercaptoethylamine, 2-aminoetha- (Fig. 1b). Taurine is excreted either in urine, or in feces in the form nethiol, 2-mercaptoethylamine, decarboxycysteine, thioethano- of bile salts [3]. -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

Exogenous Potassium
plants Article Exogenous Potassium (K+) Positively Regulates Na+/H+ Antiport System, Carbohydrate Metabolism, and Ascorbate–Glutathione Cycle in H2S-Dependent Manner in NaCl-Stressed Tomato Seedling Roots M. Nasir Khan 1,*, Soumya Mukherjee 2 , Asma A. Al-Huqail 3, Riyadh A. Basahi 1, Hayssam M. Ali 3 , Bander M. A. Al-Munqedhi 3, Manzer H. Siddiqui 3,* and Hazem M. Kalaji 4 1 Department of Biology, Faculty of Science, College of Haql, University of Tabuk, Tabuk 71491, Saudi Arabia; [email protected] 2 Department of Botany, Jangipur College, University of Kalyani, West Bengal 742213, India; [email protected] 3 Chair of Climate Change, Environmental Development and Vegetation Cover, Department of Botany and Microbiology, College of Science, King Saud University, Riyadh 11451, Saudi Arabia; [email protected] (A.A.A.-H.); [email protected] (H.M.A.); [email protected] (B.M.A.A.-M.) 4 Department of Plant Physiology, Institute of Biology, Warsaw University of Life Sciences SGGW, 159 Nowoursynowska 159, 02-776 Warsaw, Poland; [email protected] * Correspondence: [email protected] (M.N.K.); [email protected] (M.H.S.) Citation: Khan, M.N.; Mukherjee, S.; Al-Huqail, A.A.; Basahi, R.A.; Ali, Abstract: Potassium (K+) is one of the vital macronutrients required by plants for proper growth H.M.; Al-Munqedhi, B.M.A.; and blossoming harvest. In addition, K+ also plays a decisive role in promoting tolerance to various Siddiqui, M.H.; Kalaji, H.M. Exogenous Potassium (K+) Positively stresses. Under stressful conditions, plants deploy their defense system through various signaling Regulates Na+/H+ Antiport System, molecules, including hydrogen sulfide (H2S).