Downloaded from ENCODE [24, 27] Total Number of 185,297 Peaks Across All Eight Samples
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Combined Analysis of Chip-Seq and Gene Microarray Datasets Identify the E2-Mediated Genes in Erα-Dependent Manner in Osteosarcoma
ONCOLOGY REPORTS 38: 2335-2342, 2017 Combined analysis of ChIP-seq and gene microarray datasets identify the E2-mediated genes in ERα-dependent manner in osteosarcoma KANGSONG TIAN1, WEI QI1, QIAN YAN1, FEnG ZHanG1, DELEI SONG1, HaIyanG ZHanG2 and MING LV1 1Trauma Department of Orthopedics, 2Microscopic Department of Orthopedics, Zibo Central Hospital, Zibo, Shandong 255036, P.R. China Received March 14, 2017; Accepted August 11, 2017 DOI: 10.3892/or.2017.5914 Abstract. Osteosarcoma is a common bone tumor which is Introduction affected by E2, the most representative estrogen. Gene regula- tion function of E2 is highly dependent on estrogen receptor. ESR1, also known as ER-alpha or ERα, is an important The purpose of this study was to explore the gene regulation estrogen receptor (ER) and involves in the gene regula- patterns of E2 through estrogen receptor α (ESR1) in osteo- tion in various diseases and biological processes, including sarcoma based on the combined analysis of ChIP-seq and breast cancer (1,2), osteosarcoma (3) and cell growth (4). gene microarray. All of the datasets were downloaded from 17β-estradiol (E2) is one of the most representative estrogens the Gene Expression Omnibus (GEO). Differential expression responsible for the development and reproductive capability genes (DEGs) in E2 treated U2OS cells expressing ESR1 of female characteristics (5). Besides, it participates in the (U2OS-ERα) compared with those treated with vehicle were progression of many diseases, for example, E2 could regulate obtained based on R programming software. ESR1-specific the proliferation of breast cancer cells through focal adhesion binding sites (peaks) in E2 treated U2OS cells were identified and chemokine signaling pathways (6); through autophagy through MACS. -

ATRX Induction by Mutant Huntingtin Via Cdx2 Modulates Heterochromatin Condensation and Pathology in Huntington’S Disease
Cell Death and Differentiation (2012) 19, 1109–1116 & 2012 Macmillan Publishers Limited All rights reserved 1350-9047/12 www.nature.com/cdd ATRX induction by mutant huntingtin via Cdx2 modulates heterochromatin condensation and pathology in Huntington’s disease J Lee1,2, YK Hong3, GS Jeon4, YJ Hwang4, KY Kim4, KH Seong4, M-K Jung4, DJ Picketts5, NW Kowall1,2, KS Cho3 and H Ryu*,1,2,4 Aberrant chromatin remodeling is involved in the pathogenesis of Huntington’s disease (HD) but the mechanism is not known. Herein, we report that mutant huntingtin (mtHtt) induces the transcription of alpha thalassemia/mental retardation X linked (ATRX), an ATPase/helicase and SWI/SNF-like chromatin remodeling protein via Cdx-2 activation. ATRX expression was elevated in both a cell line model and transgenic model of HD, and Cdx-2 occupancy of the ATRX promoter was increased in HD. Induction of ATRX expanded the size of promyelocytic leukemia nuclear body (PML-NB) and increased trimethylation of H3K9 (H3K9me3) and condensation of pericentromeric heterochromatin, while knockdown of ATRX decreased PML-NB and H3K9me3 levels. Knockdown of ATRX/dXNP improved the hatch rate of fly embryos expressing mtHtt (Q127). ATRX/dXNP overexpression exacerbated eye degeneration of eye-specific mtHtt (Q127) expressing flies. Our findings suggest that transcriptional alteration of ATRX by mtHtt is involved in pericentromeric heterochromatin condensation and contributes to the pathogenesis of HD. Cell Death and Differentiation (2012) 19, 1109–1116; doi:10.1038/cdd.2011.196; published -

Reproductionreview
REPRODUCTIONREVIEW Forkhead transcription factors in ovarian function Nina Henriette Uhlenhaut and Mathias Treier Max Delbru¨ck Center for Molecular Medicine, Robert Ro¨ssle Straße 10, 13125 Berlin-Buch, Germany Correspondence should be addressed to N H Uhlenhaut; Email: [email protected] Abstract Since the discovery of the conserved forkhead (Fkh) DNA binding domain more than 20 years ago, members of the Fkh or forkhead box (FOX) family of transcription factors have been shown to act as important regulators of numerous developmental and homeostatic processes. The human genome contains 44 Fkh genes, several of which have recently been reported to be essential for female fertility. In this review, we highlight the roles of specific FOX proteins in ovarian folliculogenesis and present our current understanding of their molecular function. In particular, we describe what we have learned from loss-of-function studies using mouse models as well as human genetics and illustrate how different stages of folliculogenesis, both in oocytes and in somatic granulosa and theca cells, are regulated by FOXC1, FOXL2, and FOXO subfamily members. Reproduction (2011) 142 489–495 Introduction stages of folliculogenesis. This transition is marked by oocyte growth and proliferation of the adjacent granu- Female fertility depends on a delicate balance of losa cells that become cuboidal. Antral follicles are hormonal stimuli and cellular interactions, which formed when fluid-filled spaces develop between the ultimately enable conception and a successful preg- multi-layered granulosa cells. During preantral to antral nancy. Current estimates state that 10–15% of couples transition, the oocyte resumes meiosis, and after worldwide remain childless due to infertility, with stimulation by pituitary gonadotropins, FSH, and LH, genetic etiology making up a significant proportion the mature oocyte is expelled from the follicle and moves (Matzuk & Lamb 2002). -

Prospective Isolation of NKX2-1–Expressing Human Lung Progenitors Derived from Pluripotent Stem Cells
The Journal of Clinical Investigation RESEARCH ARTICLE Prospective isolation of NKX2-1–expressing human lung progenitors derived from pluripotent stem cells Finn Hawkins,1,2 Philipp Kramer,3 Anjali Jacob,1,2 Ian Driver,4 Dylan C. Thomas,1 Katherine B. McCauley,1,2 Nicholas Skvir,1 Ana M. Crane,3 Anita A. Kurmann,1,5 Anthony N. Hollenberg,5 Sinead Nguyen,1 Brandon G. Wong,6 Ahmad S. Khalil,6,7 Sarah X.L. Huang,3,8 Susan Guttentag,9 Jason R. Rock,4 John M. Shannon,10 Brian R. Davis,3 and Darrell N. Kotton1,2 2 1Center for Regenerative Medicine, and The Pulmonary Center and Department of Medicine, Boston University School of Medicine, Boston, Massachusetts, USA. 3Center for Stem Cell and Regenerative Medicine, Brown Foundation Institute of Molecular Medicine, University of Texas Health Science Center, Houston, Texas, USA. 4Department of Anatomy, UCSF, San Francisco, California, USA. 5Division of Endocrinology, Diabetes and Metabolism, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, Massachusetts, USA. 6Department of Biomedical Engineering and Biological Design Center, Boston University, Boston, Massachusetts, USA. 7Wyss Institute for Biologically Inspired Engineering, Harvard University, Boston, Massachusetts, USA. 8Columbia Center for Translational Immunology & Columbia Center for Human Development, Columbia University Medical Center, New York, New York, USA. 9Department of Pediatrics, Monroe Carell Jr. Children’s Hospital, Vanderbilt University, Nashville, Tennessee, USA. 10Division of Pulmonary Biology, Cincinnati Children’s Hospital, Cincinnati, Ohio, USA. It has been postulated that during human fetal development, all cells of the lung epithelium derive from embryonic, endodermal, NK2 homeobox 1–expressing (NKX2-1+) precursor cells. -

The Title of the Dissertation
UNIVERSITY OF CALIFORNIA SAN DIEGO Novel network-based integrated analyses of multi-omics data reveal new insights into CD8+ T cell differentiation and mouse embryogenesis A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Bioinformatics and Systems Biology by Kai Zhang Committee in charge: Professor Wei Wang, Chair Professor Pavel Arkadjevich Pevzner, Co-Chair Professor Vineet Bafna Professor Cornelis Murre Professor Bing Ren 2018 Copyright Kai Zhang, 2018 All rights reserved. The dissertation of Kai Zhang is approved, and it is accept- able in quality and form for publication on microfilm and electronically: Co-Chair Chair University of California San Diego 2018 iii EPIGRAPH The only true wisdom is in knowing you know nothing. —Socrates iv TABLE OF CONTENTS Signature Page ....................................... iii Epigraph ........................................... iv Table of Contents ...................................... v List of Figures ........................................ viii List of Tables ........................................ ix Acknowledgements ..................................... x Vita ............................................. xi Abstract of the Dissertation ................................. xii Chapter 1 General introduction ............................ 1 1.1 The applications of graph theory in bioinformatics ......... 1 1.2 Leveraging graphs to conduct integrated analyses .......... 4 1.3 References .............................. 6 Chapter 2 Systematic -
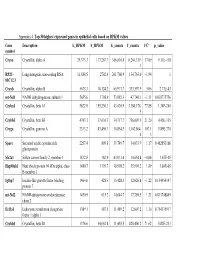
Appendix 4. Top 50 Highest Expressed Genes in Epithelial Cells Based on RPKM Values
Appendix 4. Top 50 highest expressed genes in epithelial cells based on RPKM values Gene Description E_RPKM F_RPKM E_counts F_counts FC* p_value symbol Cryaa Crystallin, alpha A 29,373.3 177,267.7 366,616.4 6,264,319. 17.09 9.11E-118 1 RP23– Long intergenic non-coding RNA 11,888.5 2702.4 261,760.9 134,763.0 −1.94 1 81C12.3 Cryab Crystallin, alpha B 5673.3 10,124.2 65,971.7 333,597.9 5.06 2.71E-43 mt-Nd1 NADH dehydrogenase, subunit 1 5655.6 1798.9 53,082.3 47,748.1 −1.11 0.838775756 Cryba1 Crystallin, beta A1 5622.0 155,230.3 43,420.9 3,380,176. 77.85 1.34E-240 5 Crybb3 Crystallin, beta B3 4743.1 37,636.3 34,717.7 736,007.9 21.20 4.45E-135 Cryga Crystallin, gamma A 2333.2 83,496.3 10,854.5 1,162,864. 107.1 5.89E-270 6 3 Sparc Secreted acidic cysteine rich 2257.4 809.8 39,749.7 34,033.9 −1.17 0.462853166 glycoprotein Slc2a1 Solute carrier family 2, member 1 1832.8 162.9 43,031.4 10,654.8 −4.04 1.67E-05 Hsp90ab1 Heat shock protein 90 kDa alpha, class 1480.7 1139.7 18,998.2 35,901.2 1.89 3.84E-05 B member 1 Igfbp7 Insulin-like growth factor binding 1464.6 428.3 15,428.3 12,626.8 −1.22 0.154954147 protein 7 mt-Nd2 NADH-ubiquinone oxidoreductase 1450.9 615.2 14,644.7 17,789.5 1.21 0.833748849 chain 2 Eef1a1 Eukaryotic translation elongation 1389.1 587.5 11,489.2 12,607.2 1.10 0.754135917 factor 1 alpha 1 Crybb1 Crystallin, beta B1 1376.6 34,662.8 11,455.5 820,406.2 71.62 5.82E-233 Htra3 HtrA serine peptidase 3 1338.6 162.0 23,197.6 6433.9 −3.61 3.93E-05 Gnb2l1 Guanine nucleotide-binding protein 1293.3 670.1 14,495.1 21,652.1 1.49 0.001685952 -

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

Integrated and Functional Genomic Approaches to Elucidate Differential Genetic Dependencies in Melanoma
Integrated and Functional Genomic Approaches to Elucidate Differential Genetic Dependencies in Melanoma The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Wong, Terence. 2018. Integrated and Functional Genomic Approaches to Elucidate Differential Genetic Dependencies in Melanoma. Doctoral dissertation, Harvard University, Graduate School of Arts & Sciences. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:42014990 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA Integrated and Functional Genomic Approaches to Elucidate Differential Genetic Dependencies in Melanoma A dissertation presented by Terence Cheng Wong to The Division of Medical Sciences in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the subject of Biological and Biomedical Sciences Harvard University Cambridge, Massachusetts November 2017 © 2017 Terence Cheng Wong All rights reserved. Dissertation Advisor: Levi Garraway Terence Cheng Wong Integrated and Functional Genomic Approaches to Elucidate Differential Genetic Dependencies in Melanoma ABSTRACT Genomic characterization of human cancers over the past decade has generated comprehensive catalogues of genetic alterations in cancer genomes. Many of these genetic events result in molecular or cellular changes that drive cancer cell phenotypes. In melanoma, a majority of tumors harbor mutations in the BRAF gene, leading to activation of the MAPK pathway and tumor initiation. The development and use of drugs that target the mutant BRAF protein and the MAPK pathway have produced significant clinical benefit in melanoma patients. -

C-Myc Regulates Cell Proliferation During Lens Development
c-myc Regulates Cell Proliferation during Lens Development Gabriel R. Cavalheiro., Gabriel E. Matos-Rodrigues., Anielle L. Gomes, Paulo M. G. Rodrigues, Rodrigo A. P. Martins* Programa de Biologia Celular e do Desenvolvimento, Instituto de Cieˆncias Biome´dicas, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Rio de Janeiro, Brazil Abstract Myc protooncogenes play important roles in the regulation of cell proliferation, growth, differentiation and survival during development. In various developing organs, c-myc has been shown to control the expression of cell cycle regulators and its misregulated expression is detected in many human tumors. Here, we show that c-myc gene (Myc) is highly expressed in developing mouse lens. Targeted deletion of c-myc gene from head surface ectoderm dramatically impaired ocular organogenesis, resulting in severe microphtalmia, defective anterior segment development, formation of a lens stalk and/or aphakia. In particular, lenses lacking c-myc presented thinner epithelial cell layer and growth impairment that was detectable soon after its inactivation. Defective development of c-myc-null lens was not caused by increased cell death of lens progenitor cells. Instead, c-myc loss reduced cell proliferation, what was associated with an ectopic expression of Prox1 and p27Kip1 proteins within epithelial cells. Interestingly, a sharp decrease in the expression of the forkhead box transcription factor Foxe3 was also observed following c-myc inactivation. These data represent the first description of the physiological roles played by a Myc family member in mouse lens development. Our findings support the conclusion that c- myc regulates the proliferation of lens epithelial cells in vivo and may, directly or indirectly, modulate the expression of classical cell cycle regulators in developing mouse lens. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

The E–Id Protein Axis Modulates the Activities of the PI3K–AKT–Mtorc1
Downloaded from genesdev.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press The E–Id protein axis modulates the activities of the PI3K–AKT–mTORC1– Hif1a and c-myc/p19Arf pathways to suppress innate variant TFH cell development, thymocyte expansion, and lymphomagenesis Masaki Miyazaki,1,8 Kazuko Miyazaki,1,8 Shuwen Chen,1 Vivek Chandra,1 Keisuke Wagatsuma,2 Yasutoshi Agata,2 Hans-Reimer Rodewald,3 Rintaro Saito,4 Aaron N. Chang,5 Nissi Varki,6 Hiroshi Kawamoto,7 and Cornelis Murre1 1Department of Molecular Biology, University of California at San Diego, La Jolla, California 92093, USA; 2Department of Biochemistry and Molecular Biology, Shiga University of Medical School, Shiga 520-2192, Japan; 3Division of Cellular Immunology, German Cancer Research Center, D-69120 Heidelberg, Germany; 4Department of Medicine, University of California at San Diego, La Jolla, California 92093, USA; 5Center for Computational Biology, Institute for Genomic Medicine, University of California at San Diego, La Jolla, California 92093, USA; 6Department of Pathology, University of California at San Diego, La Jolla, California 92093, USA; 7Department of Immunology, Institute for Frontier Medical Sciences, Kyoto University, Kyoto 606-8507, Japan It is now well established that the E and Id protein axis regulates multiple steps in lymphocyte development. However, it remains unknown how E and Id proteins mechanistically enforce and maintain the naı¨ve T-cell fate. Here we show that Id2 and Id3 suppressed the development and expansion of innate variant follicular helper T (TFH) cells. Innate variant TFH cells required major histocompatibility complex (MHC) class I-like signaling and were associated with germinal center B cells. -

Supp Table 1.Pdf
Upregulated genes in Hdac8 null cranial neural crest cells fold change Gene Symbol Gene Title 134.39 Stmn4 stathmin-like 4 46.05 Lhx1 LIM homeobox protein 1 31.45 Lect2 leukocyte cell-derived chemotaxin 2 31.09 Zfp108 zinc finger protein 108 27.74 0710007G10Rik RIKEN cDNA 0710007G10 gene 26.31 1700019O17Rik RIKEN cDNA 1700019O17 gene 25.72 Cyb561 Cytochrome b-561 25.35 Tsc22d1 TSC22 domain family, member 1 25.27 4921513I08Rik RIKEN cDNA 4921513I08 gene 24.58 Ofa oncofetal antigen 24.47 B230112I24Rik RIKEN cDNA B230112I24 gene 23.86 Uty ubiquitously transcribed tetratricopeptide repeat gene, Y chromosome 22.84 D8Ertd268e DNA segment, Chr 8, ERATO Doi 268, expressed 19.78 Dag1 Dystroglycan 1 19.74 Pkn1 protein kinase N1 18.64 Cts8 cathepsin 8 18.23 1500012D20Rik RIKEN cDNA 1500012D20 gene 18.09 Slc43a2 solute carrier family 43, member 2 17.17 Pcm1 Pericentriolar material 1 17.17 Prg2 proteoglycan 2, bone marrow 17.11 LOC671579 hypothetical protein LOC671579 17.11 Slco1a5 solute carrier organic anion transporter family, member 1a5 17.02 Fbxl7 F-box and leucine-rich repeat protein 7 17.02 Kcns2 K+ voltage-gated channel, subfamily S, 2 16.93 AW493845 Expressed sequence AW493845 16.12 1600014K23Rik RIKEN cDNA 1600014K23 gene 15.71 Cst8 cystatin 8 (cystatin-related epididymal spermatogenic) 15.68 4922502D21Rik RIKEN cDNA 4922502D21 gene 15.32 2810011L19Rik RIKEN cDNA 2810011L19 gene 15.08 Btbd9 BTB (POZ) domain containing 9 14.77 Hoxa11os homeo box A11, opposite strand transcript 14.74 Obp1a odorant binding protein Ia 14.72 ORF28 open reading