Kinase Inhibitors
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Supplementary Materials
1 Supplementary Materials: Supplemental Figure 1. Gene expression profiles of kidneys in the Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice. (A) A heat map of microarray data show the genes that significantly changed up to 2 fold compared between Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice (N=4 mice per group; p<0.05). Data show in log2 (sample/wild-type). 2 Supplemental Figure 2. Sting signaling is essential for immuno-phenotypes of the Fcgr2b-/-lupus mice. (A-C) Flow cytometry analysis of splenocytes isolated from wild-type, Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice at the age of 6-7 months (N= 13-14 per group). Data shown in the percentage of (A) CD4+ ICOS+ cells, (B) B220+ I-Ab+ cells and (C) CD138+ cells. Data show as mean ± SEM (*p < 0.05, **p<0.01 and ***p<0.001). 3 Supplemental Figure 3. Phenotypes of Sting activated dendritic cells. (A) Representative of western blot analysis from immunoprecipitation with Sting of Fcgr2b-/- mice (N= 4). The band was shown in STING protein of activated BMDC with DMXAA at 0, 3 and 6 hr. and phosphorylation of STING at Ser357. (B) Mass spectra of phosphorylation of STING at Ser357 of activated BMDC from Fcgr2b-/- mice after stimulated with DMXAA for 3 hour and followed by immunoprecipitation with STING. (C) Sting-activated BMDC were co-cultured with LYN inhibitor PP2 and analyzed by flow cytometry, which showed the mean fluorescence intensity (MFI) of IAb expressing DC (N = 3 mice per group). 4 Supplemental Table 1. Lists of up and down of regulated proteins Accession No. -
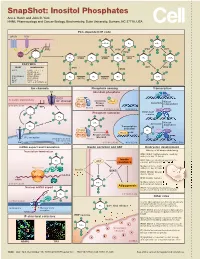
Snapshot: Inositol Phosphates Ace J
SnapShot: Inositol Phosphates Ace J. Hatch and John D. York HHMI, Pharmacology and Cancer Biology, Biochemistry, Duke University, Durham, NC 27710, USA PLC-dependent IP code GPCR RTK O O O O O O 5-PP-IP4 IP4 5-IP7 O O O O O O PIP2 O IP6K O IP6K O VIP1 O O O 2 O ITPK1 O 13 O PLC 2 O O O O O O O O 4 6 13 O 5 IP3 IPMK IP4 IPMK IP5 IPK1 IP6 1,5-IP8 4 6 O O 5 O O O O O O O O ENZYMES O O O O O O YEAST MAMMALIAN IP3K VIP1 IP6K IPMK PLC1 PLCβ, γ, δ, ε, ζ, η O - IP3KA, B, C - ITPK1 (IP56K) O O O O O O O O IPK2(ARG82) IPMK (IPK2) IP4 IP3 IP4 1-IP7 IPK1 IPK1 (IP5K) INPP5 ITPK1 KCS1 IP6K1, 2, 3 O O O O O O VIP1 VIP1, 2 (PPIP5K1, 2) O O Ion channels Phosphate sensing Transcription Cl- Abundant phosphate MCM1 ARG80 CIC3 P PLASMA MEMBRANE - Pho80 Cl channel Pho4 Kinase Kinase Assembly Pho85 independent CYTOPLASM activity 2 O PIP2 Pho81 13 CYTOPLASM NUCLEUS IPK2 ARG81 4 6 Phosphate starvation MCM1-ArgR O 5 O complex O O IP4 O O O O O O O 1-IP7 Kinase Activation dependent IP3 O O Transcription O O O activated Pho80 IP4 O X Pho4 O O Pho85 Kinase activity IP receptor blocked O 3 ENDOPLASMIC Pho81 RETICULUM Ca2+ CYTOPLASM NUCLEUS NUCLEUS mRNA export and translation Insulin secretion and AKT Embryonic development Translation termination Effects of IP kinase deficiency O IPMK (IPK2): Multiple defects, death by embryonic day 10 (mice) O O Insulin IPK1: Cillia are shortened and immotile IP6 AKT resistance causing patterning defects (zebrash) O O Multiple defects, death by Ribosome O embryonic day 8.5 (mice) GleI eRF1 Insulin GSK3β Dbp5 ITPK1 (IP56K): Neural tube -

Novocib Nucleoside Kinases
PRECICE ® Services Information sheet Nucleoside kinases, rate-limiting step of nucleoside analogues activation Nucleoside analogues have proven to be a highly successful class of anti-cancer and anti-viral drugs. The therapeutic efficacy of nucleoside analogues is dependent of their intracellular phosphorylation. Two cellular nucleoside kinases, deoxycytidine kinase (dCK) and UMP-CMP kinase (CMK) are critical for phosphorylation of cytidine analogues. These kinases provide two first steps of activation of highly effective anti-cancer and anti-viral drugs, such as 1-β-D- arabinofuranosylcytosine (araC, aracytidine ), 2’,2’difluorodeoxycytidine (dFdC, gemcitabine ), β-D-2’3’- dideoxycytidine (ddC). Both kinases phosphorylate unnatural L-nucleosides (e.g., β-L-2’3’-dideoxy-3’thiacytidine, L- SSdC, 3-TC or lamividune ). Kinetic constants of araC, dFdC and 3TC phosphorylation by recombinant dCK and UMP-CMPK have been published. The comparison of phosphorylation properties of new nucleoside analogues with those of known drugs provides the rational basis for selection of analogues of better therapeutic potential. To characterize the phosphorylation properties of new nucleoside analogues, Novo CIB has developed human recombinant dCK and human recombinant CMK nucleoside phosphorylation assays. As shown in Table 1, CMK assay must be performed with monophosphate forms of nucleoside analogues and requires preliminary phosphorylation of nucleoside analogues and their purification. To circumvent this time-consuming step, NovoCIB has developed a coupled dCK-CMK nucleoside phosphorylation assay that delivers in one step the critical information on both dCK and CMK substrate properties of nucleoside analogue. Ribavirin (1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide) is a purine nucleoside analogue with a broad-spectrum antiviral activity. -

Ribonucleotide Reductase Messenger RNA Expression and Survival in Gemcitabine/Cisplatin-Treated Advanced Non-Small Cell Lung Cancer Patients
1318 Vol. 10, 1318–1325, February 15, 2004 Clinical Cancer Research Ribonucleotide Reductase Messenger RNA Expression and Survival in Gemcitabine/Cisplatin-Treated Advanced Non-Small Cell Lung Cancer Patients Rafael Rosell,1 Kathleen D. Danenberg,2 from bronchoscopy by real-time quantitative reverse tran- Vincente Alberola,3 Gerold Bepler,4 scription-PCR. Results were correlated with survival using Jose Javier Sanchez,5 Carlos Camps,6 the Kaplan-Meier method. 7 8 Results: A total of 100 patients were assessed. There was Mariano Provencio, Dolores Isla, a strong correlation between RRM1 and ERCC1 mRNA 1 9 P < 0.001). In the ;0.410 ؍ Miquel Taron, Pilar Diz, and expression levels (Spearman r Angel Artal10 on behalf of the Spanish Lung gemcitabine/cisplatin arm, patients with low RRM1 mRNA Cancer Group expression levels had significantly longer median survival 1Medical Oncology Service, Institut Catala`d’Oncologia, Hospital than those with high levels [13.7 versus 3.6 months; 95% -Me .[0.009 ؍ Germans Trias i Pujol, Barcelona, Spain; 2Response Genetics Inc, confidence interval (CI), 9.6–17.8 months; P Los Angeles, California; 3Medical Oncology Service, Hospital Arnau dian survival was also significantly longer among patients 4 de Vilanova, Valencia, Spain; Thoracic Oncology Program, H. Lee with low mRNA expression levels of both RRM1 and ERCC1 Moffitt Cancer Center and Research Institute, Tampa, Florida; 5 (not reached), than among those with high levels of both ؍ ,Facultat de Medicina, Autonomous University of Madrid, Madrid Spain; 6Medical Oncology Service, Hospital General de Valencia, genes (6.8 months; 95% CI, 2.6–11.1 months; P 0.016). -

Supplementary File 2A Revised
Supplementary file 2A. Differentially expressed genes in aldosteronomas compared to all other samples, ranked according to statistical significance. Missing values were not allowed in aldosteronomas, but to a maximum of five in the other samples. Acc UGCluster Name Symbol log Fold Change P - Value Adj. P-Value B R99527 Hs.8162 Hypothetical protein MGC39372 MGC39372 2,17 6,3E-09 5,1E-05 10,2 AA398335 Hs.10414 Kelch domain containing 8A KLHDC8A 2,26 1,2E-08 5,1E-05 9,56 AA441933 Hs.519075 Leiomodin 1 (smooth muscle) LMOD1 2,33 1,3E-08 5,1E-05 9,54 AA630120 Hs.78781 Vascular endothelial growth factor B VEGFB 1,24 1,1E-07 2,9E-04 7,59 R07846 Data not found 3,71 1,2E-07 2,9E-04 7,49 W92795 Hs.434386 Hypothetical protein LOC201229 LOC201229 1,55 2,0E-07 4,0E-04 7,03 AA454564 Hs.323396 Family with sequence similarity 54, member B FAM54B 1,25 3,0E-07 5,2E-04 6,65 AA775249 Hs.513633 G protein-coupled receptor 56 GPR56 -1,63 4,3E-07 6,4E-04 6,33 AA012822 Hs.713814 Oxysterol bining protein OSBP 1,35 5,3E-07 7,1E-04 6,14 R45592 Hs.655271 Regulating synaptic membrane exocytosis 2 RIMS2 2,51 5,9E-07 7,1E-04 6,04 AA282936 Hs.240 M-phase phosphoprotein 1 MPHOSPH -1,40 8,1E-07 8,9E-04 5,74 N34945 Hs.234898 Acetyl-Coenzyme A carboxylase beta ACACB 0,87 9,7E-07 9,8E-04 5,58 R07322 Hs.464137 Acyl-Coenzyme A oxidase 1, palmitoyl ACOX1 0,82 1,3E-06 1,2E-03 5,35 R77144 Hs.488835 Transmembrane protein 120A TMEM120A 1,55 1,7E-06 1,4E-03 5,07 H68542 Hs.420009 Transcribed locus 1,07 1,7E-06 1,4E-03 5,06 AA410184 Hs.696454 PBX/knotted 1 homeobox 2 PKNOX2 1,78 2,0E-06 -

Ceramide Kinase Is Upregulated in Metastatic Breast Cancer Cells and Contributes to Migration and Invasion by Activation of PI 3-Kinase and Akt
International Journal of Molecular Sciences Article Ceramide Kinase Is Upregulated in Metastatic Breast Cancer Cells and Contributes to Migration and Invasion by Activation of PI 3-Kinase and Akt 1,2, 1, 2 2 Stephanie Schwalm y, Martin Erhardt y, Isolde Römer , Josef Pfeilschifter , Uwe Zangemeister-Wittke 1,* and Andrea Huwiler 1,* 1 Institute of Pharmacology, University of Bern, Inselspital, INO-F, CH-3010 Bern, Switzerland; [email protected] (S.S.); [email protected] (M.E.) 2 Institute of General Pharmacology and Toxicology, University Hospital Frankfurt am Main, Goethe-University, Theodor-Stern Kai 7, D-60590 Frankfurt am Main, Germany; [email protected] (I.R.); [email protected] (J.P.) * Correspondence: [email protected] (U.Z.-W.); [email protected] (A.H.); Tel.: +41-31-6323214 (A.H.) These authors contributed equally to this work. y Received: 29 November 2019; Accepted: 12 February 2020; Published: 19 February 2020 Abstract: Ceramide kinase (CerK) is a lipid kinase that converts the proapoptotic ceramide to ceramide 1-phosphate, which has been proposed to have pro-malignant properties and regulate cell responses such as proliferation, migration, and inflammation. We used the parental human breast cancer cell line MDA-MB-231 and two single cell progenies derived from lung and bone metastasis upon injection of the parental cells into immuno-deficient mice. The lung and the bone metastatic cell lines showed a marked upregulation of CerK mRNA and activity when compared to the parental cell line. The metastatic cells also had increased migratory and invasive activity, which was dose-dependently reduced by the selective CerK inhibitor NVP-231. -

The KMT1A-GATA3-STAT3 Circuit Is a Novel Self-Renewal Signaling of Human Bladder Cancer Stem Cells Zhao Yang1, Luyun He2,3, Kais
The KMT1A-GATA3-STAT3 circuit is a novel self-renewal signaling of human bladder cancer stem cells Zhao Yang1, Luyun He2,3, Kaisu Lin4, Yun Zhang1, Aihua Deng1, Yong Liang1, Chong Li2, 5, & Tingyi Wen1, 6, 1CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China 2Core Facility for Protein Research, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China 3CAS Key Laboratory of Infection and Immunity, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China 4Department of Oncology, the Second Affiliated Hospital of Soochow University, Suzhou 215000, China 5Beijing Jianlan Institute of Medicine, Beijing 100190, China 6Savaid Medical School, University of Chinese Academy of Sciences, Beijing 100049, China Correspondence author: Tingyi Wen, e-mail: [email protected] Chong Li, e-mail: [email protected] Supplementary Figure S1. Isolation of human bladder cancer stem cells. BCMab1 and CD44 were used to isolate bladder cancer stem cells (BCSCs: BCMab1+CD44+) and bladder cancer non-stem cells (BCNSCs: BCMab1-CD44-) from EJ, samples #1 and #2 by flow cytometry. Supplementary Figure S2. Gene ontology analysis of downregulated genes of human BCSCs. (A) Pathway enrichment of 103 downregulated genes in BCSCs. (B) The seven downregulated genes in BCSCs participating in centromeric heterochromatin, mRNA-3’-UTR binding and translation regulator activity signaling pathways were validated by qRT-PCR. Data are presented as mean ± SD. P < 0.05; P < 0.01. Supplementary Figure S3. The expression of KMT1A is higher in human BC than that in peri-tumor tissues. (A) The expression of KMT1A was higher in BC samples than that in peri-tumors as assessed by immunohistochemistry, Scale bar = 50 m. -

Inhibition of Spinach Phosphoribulokinase by DL-Glyceraldehyde by ANTONI R
Biochem. J. (1976) 153, 613-619 613 Printed in Great Britain Inhibition of Spinach Phosphoribulokinase by DL-Glyceraldehyde By ANTONI R. SLABAS and DAVID A. WALKER Department ofBotany, University ofSheffield, Sheffield S1O 2TN, U.K. (Received 10 September 1975) Spinach chloroplast phosphoribulokinase is inhibited by DL-glyceraldehyde. The in- hibition is non-competitive with respect to ribulose 5-phosphate (Ki 19mM) and ATP (Ki 20mM). The inhibition is discussed in relation to a previously reported inhibition of CO2 assimilation in intact and envelope-free chloroplasts by DL-glyceraldehyde. It is concluded that the inhibition of phosphoribulokinase is insufficient to account for the inhibition, by DL-glyceraldehyde, of 02 evolution with ribose 5-phosphate as substrate and that a further site of inhibition is also present in this system. DL-Glyceraldehyde inhibits photosynthetic carbon glycylglycine buffer, pH7.4, in a total volume of assiniilation by intact chloroplasts and by the re- 13 ml. The reaction was terminated by the addition constituted chloroplast system (Stokes & Walker, of 2ml of 50 % (w/v) trichloroacetic acid and the pH 1972). The inhibition is most pronounced with intact was adjusted to pH8.0 with KOH. After the addition chloroplasts, a concentration of 1OmM being sufficient of Sml of 1.OM-barium acetate the solution was to suppress 02 evolution completely. It is important centrifuged and the precipitate discarded after because DL-glyceraldehyde is the only known inhibi- washing with Sml of water. Cold ethanol (4vol.) was tor of photosynthesis that is entirely without detect- added to the combined supernatants (total volume able effect on photophosphorylation or photo- 25.4ml); the new precipitate was recovered by synthetic electron transport. -

Swiveling Domain Mechanism in Pyruvate Phosphate Dikinase†,‡ Kap Lim,§ Randy J
Biochemistry 2007, 46, 14845-14853 14845 Swiveling Domain Mechanism in Pyruvate Phosphate Dikinase†,‡ Kap Lim,§ Randy J. Read,| Celia C. H. Chen,§ Aleksandra Tempczyk,§ Min Wei,⊥ Dongmei Ye,⊥ Chun Wu,⊥ Debra Dunaway-Mariano,⊥ and Osnat Herzberg*,§ Center for AdVanced Research in Biotechnology, UniVersity of Maryland Biotechnology Institute, RockVille, Maryland 20850, Department of Haematology, Cambridge Institute for Medical Research, UniVersity of Cambridge, Cambridge, United Kingdom, and Department of Chemistry, UniVersity of New Mexico, Albuquerque, New Mexico ReceiVed September 10, 2007; ReVised Manuscript ReceiVed October 17, 2007 ABSTRACT: Pyruvate phosphate dikinase (PPDK) catalyzes the reversible conversion of phosphoenolpyruvate (PEP), AMP, and Pi to pyruvate and ATP. The enzyme contains two remotely located reaction centers: the nucleotide partial reaction takes place at the N-terminal domain, and the PEP/pyruvate partial reaction takes place at the C-terminal domain. A central domain, tethered to the N- and C-terminal domains by two closely associated linkers, contains a phosphorylatable histidine residue (His455). The molecular architecture suggests a swiveling domain mechanism that shuttles a phosphoryl group between the two reaction centers. In an early structure of PPDK from Clostridium symbiosum, the His445-containing domain (His domain) was positioned close to the nucleotide binding domain and did not contact the PEP/pyruvate- binding domain. Here, we present the crystal structure of a second conformational state of C. symbiosum PPDK with the His domain adjacent to the PEP-binding domain. The structure was obtained by producing a three-residue mutant protein (R219E/E271R/S262D) that introduces repulsion between the His and nucleotide-binding domains but preserves viable interactions with the PEP/pyruvate-binding domain. -

Ceramides: Nutrient Signals That Drive Hepatosteatosis
J Lipid Atheroscler. 2020 Jan;9(1):50-65 Journal of https://doi.org/10.12997/jla.2020.9.1.50 Lipid and pISSN 2287-2892·eISSN 2288-2561 Atherosclerosis Review Ceramides: Nutrient Signals that Drive Hepatosteatosis Scott A. Summers Department of Nutrition and Integrative Physiology, University of Utah, Salt Lake City, UT, USA Received: Sep 24, 2019 ABSTRACT Revised: Nov 4, 2019 Accepted: Nov 10, 2019 Ceramides are minor components of the hepatic lipidome that have major effects on liver Correspondence to function. These products of lipid and protein metabolism accumulate when the energy needs Scott A. Summers of the hepatocyte have been met and its storage capacity is full, such that free fatty acids start Department of Nutrition and Integrative to couple to the sphingoid backbone rather than the glycerol moiety that is the scaffold for Physiology, University of Utah, 15N 2030E, Salt Lake City, UT 84112, USA. glycerolipids (e.g., triglycerides) or the carnitine moiety that shunts them into mitochondria. E-mail: [email protected] As ceramides accrue, they initiate actions that protect cells from acute increases in detergent- like fatty acids; for example, they alter cellular substrate preference from glucose to lipids Copyright © 2020 The Korean Society of Lipid and they enhance triglyceride storage. When prolonged, these ceramide actions cause insulin and Atherosclerosis. This is an Open Access article distributed resistance and hepatic steatosis, 2 of the underlying drivers of cardiometabolic diseases. under the terms of the Creative Commons Herein the author discusses the mechanisms linking ceramides to the development of insulin Attribution Non-Commercial License (https:// resistance, hepatosteatosis and resultant cardiometabolic disorders. -

Glucokinase September 15, 2005
Glucokinase September 15, 2005 Summary Glucokinase phosphorylates glucose into glucose 6-phosphate using ATP as the phosphate group donor, according to the reaction glucose + ATP Æ glucose 6-phosphate + ADP This protocol describes an indirect assay to determine the activity of glucokinase. Glucose 6- phosphate formed by glucokinase is measured by the formation of NADPH in presence of glucose 6-phosphate dehydrogenase. Solutions Required 1. 100 mM tris HCl pH 7.4 Stock solution could be used 2. 100 mM MgSO4·7H2O Stock solution could be used 3. 10 mM glucose Must be prepared fresh 4. 20 mM ATP Must be prepared fresh 5. 10 mM NADP Must be prepared fresh 6. a solution containing 10 U of glucose 6-phosphate dehydrogenase per 50 μL in tris HCl pH 7.4 buffer. This solution is prepared by mixing 40 μL of a 5000 U/mL solution (Sigma G8529) with 960 μL tris buffer. Preparation of Cell Extract Follow general protocol described in Preparation of Cell Extract. The cell extract should be suspended in tris buffer after pelletization. Spectrophotometer Turn on the ultraviolet bulb on the spectrophotometer (Beckman DU50) and wait 10 minutes for warm-up. Select the kinetics-time window on the instrument. Load the method "A:/nadh" ". These methods each have a run-time of 120 s, a temperature of 37°C, a wavelength of 340 nm and use 2 autosamplers. Procedure 1. For each assay, prepare the two cocktails shown in the following table into two separate quartz cuvettes. Keep them on ice. Volume (μL) added to: Solution Control Experimental tris 500 500 DI H2O 200 100 MgSO4 100 100 glucose 0 100 ATP 50 50 NADP 50 50 G6P dehydrogenase 50 50 2.