Inhibition of Spinach Phosphoribulokinase by DL-Glyceraldehyde by ANTONI R
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Indications for a Central Role of Hexokinase Activity in Natural Variation of Heat Acclimation in Arabidopsis Thaliana
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 14 June 2020 doi:10.20944/preprints202006.0169.v1 Article Indications for a central role of hexokinase activity in natural variation of heat acclimation in Arabidopsis thaliana Vasil Atanasov §, Lisa Fürtauer § and Thomas Nägele * LMU Munich, Plant Evolutionary Cell Biology, Großhaderner Str. 2-4, 82152 Planegg, Germany § Authors contributed equally * Correspondence: [email protected] Abstract: Diurnal and seasonal changes of abiotic environmental factors shape plant performance and distribution. Changes of growth temperature and light intensity may vary significantly on a diurnal, but also on a weekly or seasonal scale. Hence, acclimation to a changing temperature and light regime is essential for plant survival and propagation. In the present study, we analyzed photosynthetic CO2 assimilation and metabolic regulation of the central carbohydrate metabolism in two natural accessions of Arabidopsis thaliana originating from Russia and south Italy during exposure to heat and a combination of heat and high light. Our findings indicate that it is hardly possible to predict photosynthetic capacities to fix CO2 under combined stress from single stress experiments. Further, capacities of hexose phosphorylation were found to be significantly lower in the Italian than in the Russian accession which could explain an inverted sucrose-to-hexose ratio. Together with the finding of significantly stronger accumulation of anthocyanins under heat/high light these observations indicate a central role of hexokinase activity in stabilization of photosynthetic capacities within a changing environment. Keywords: photosynthesis; carbohydrate metabolism; hexokinase; heat acclimation; environmental changes; natural variation; high light; combined stress. 1. Introduction Changes of growth temperature and light intensity broadly affect plant molecular, physiological and developmental processes. -

METABOLIC EVOLUTION in GALDIERIA SULPHURARIA By
METABOLIC EVOLUTION IN GALDIERIA SULPHURARIA By CHAD M. TERNES Bachelor of Science in Botany Oklahoma State University Stillwater, Oklahoma 2009 Submitted to the Faculty of the Graduate College of the Oklahoma State University in partial fulfillment of the requirements for the Degree of DOCTOR OF PHILOSOPHY May, 2015 METABOLIC EVOLUTION IN GALDIERIA SUPHURARIA Dissertation Approved: Dr. Gerald Schoenknecht Dissertation Adviser Dr. David Meinke Dr. Andrew Doust Dr. Patricia Canaan ii Name: CHAD M. TERNES Date of Degree: MAY, 2015 Title of Study: METABOLIC EVOLUTION IN GALDIERIA SULPHURARIA Major Field: PLANT SCIENCE Abstract: The thermoacidophilic, unicellular, red alga Galdieria sulphuraria possesses characteristics, including salt and heavy metal tolerance, unsurpassed by any other alga. Like most plastid bearing eukaryotes, G. sulphuraria can grow photoautotrophically. Additionally, it can also grow solely as a heterotroph, which results in the cessation of photosynthetic pigment biosynthesis. The ability to grow heterotrophically is likely correlated with G. sulphuraria ’s broad capacity for carbon metabolism, which rivals that of fungi. Annotation of the metabolic pathways encoded by the genome of G. sulphuraria revealed several pathways that are uncharacteristic for plants and algae, even red algae. Phylogenetic analyses of the enzymes underlying the metabolic pathways suggest multiple instances of horizontal gene transfer, in addition to endosymbiotic gene transfer and conservation through ancestry. Although some metabolic pathways as a whole appear to be retained through ancestry, genes encoding individual enzymes within a pathway were substituted by genes that were acquired horizontally from other domains of life. Thus, metabolic pathways in G. sulphuraria appear to be composed of a ‘metabolic patchwork’, underscored by a mosaic of genes resulting from multiple evolutionary processes. -

Isozymes of Pyruvate Kinase in Liver and Hepatomas of the Rat1
[CANCER RESEARCH 34, 1439-1446, June 1974] Isozymes of Pyruvate Kinase in Liver and Hepatomas of the Rat1 Francis A. Farina,2 Jennie B. Shatton, Harold P. Morris, and Sidney Weinhouse The Fels Research Institute and the Department of Biochemistry, Temple University School oj Medicine, Philadelphia. Pennsylvania IV140 (F. A. F., J. B. S.. S. W.\, and the Department of Biochemistry. Howard University School of Medicine. Washington. D. C. 20001 [H. P. M .\ SUMMARY 23, 32, 41, 57). These alterations involve the replacement of those isozymes that are under dietary and hormonal control Pyruvate kinase (PK) (EC 2.7.1.40) isozymes were assayed by the host, and that have important metabolic functions in in normal rat liver and a series of transplantable rat the adult differentiated liver by other isozymes which are hepatomas ranging widely in growth rate and degree of normally either low in, or absent from, the adult tissue. As differentiation, with the use of gradient elution by chloride part of an ongoing study of this phenomenon, we have ion from columns of DEAE-cellulose. In agreement with examined the alteration of PK3 (EC 2.7.1.40) isozymes in other studies, three noninterconvertible forms were found in the Morris hepatomas (30. 31), a series of chemically rat tissues: isozyme I, the major form in adult rat liver: induced, transplantable rat hepatomas ranging widely in isozyme II, the sole form in heart and skeletal muscle: and growth rate and degree of differentiation. This enzyme isozyme III, the sole form in poorly differentiated hepato occupies a key position in the metabolism of cells and, as we mas, the major form in normal kidney and lung, and the pointed out previously (27, 28. -

Pyruvate Kinase: Function, Regulation and Role in Cancer
Pyruvate kinase: Function, regulation and role in cancer The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Israelsen, William J., and Matthew G. Vander Heiden. “Pyruvate Kinase: Function, Regulation and Role in Cancer.” Seminars in Cell & Developmental Biology 43 (2015): 43–51. As Published http://dx.doi.org/10.1016/j.semcdb.2015.08.004 Publisher Elsevier Version Author's final manuscript Citable link http://hdl.handle.net/1721.1/105833 Terms of Use Creative Commons Attribution-NonCommercial-NoDerivs License Detailed Terms http://creativecommons.org/licenses/by-nc-nd/4.0/ HHS Public Access Author manuscript Author Manuscript Author ManuscriptSemin Cell Author Manuscript Dev Biol. Author Author Manuscript manuscript; available in PMC 2016 August 13. Published in final edited form as: Semin Cell Dev Biol. 2015 July ; 43: 43–51. doi:10.1016/j.semcdb.2015.08.004. Pyruvate kinase: function, regulation and role in cancer William J. Israelsena,1,* and Matthew G. Vander Heidena,b,* aKoch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02139, USA bDepartment of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02115, USA Abstract Pyruvate kinase is an enzyme that catalyzes the conversion of phosphoenolpyruvate and ADP to pyruvate and ATP in glycolysis and plays a role in regulating cell metabolism. There are four mammalian pyruvate kinase isoforms with unique tissue expression patterns and regulatory properties. The M2 isoform of pyruvate kinase (PKM2) supports anabolic metabolism and is expressed both in cancer and normal tissue. The enzymatic activity of PKM2 is allosterically regulated by both intracellular signaling pathways and metabolites; PKM2 thus integrates signaling and metabolic inputs to modulate glucose metabolism according to the needs of the cell. -
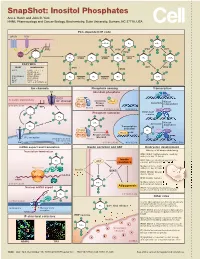
Snapshot: Inositol Phosphates Ace J
SnapShot: Inositol Phosphates Ace J. Hatch and John D. York HHMI, Pharmacology and Cancer Biology, Biochemistry, Duke University, Durham, NC 27710, USA PLC-dependent IP code GPCR RTK O O O O O O 5-PP-IP4 IP4 5-IP7 O O O O O O PIP2 O IP6K O IP6K O VIP1 O O O 2 O ITPK1 O 13 O PLC 2 O O O O O O O O 4 6 13 O 5 IP3 IPMK IP4 IPMK IP5 IPK1 IP6 1,5-IP8 4 6 O O 5 O O O O O O O O ENZYMES O O O O O O YEAST MAMMALIAN IP3K VIP1 IP6K IPMK PLC1 PLCβ, γ, δ, ε, ζ, η O - IP3KA, B, C - ITPK1 (IP56K) O O O O O O O O IPK2(ARG82) IPMK (IPK2) IP4 IP3 IP4 1-IP7 IPK1 IPK1 (IP5K) INPP5 ITPK1 KCS1 IP6K1, 2, 3 O O O O O O VIP1 VIP1, 2 (PPIP5K1, 2) O O Ion channels Phosphate sensing Transcription Cl- Abundant phosphate MCM1 ARG80 CIC3 P PLASMA MEMBRANE - Pho80 Cl channel Pho4 Kinase Kinase Assembly Pho85 independent CYTOPLASM activity 2 O PIP2 Pho81 13 CYTOPLASM NUCLEUS IPK2 ARG81 4 6 Phosphate starvation MCM1-ArgR O 5 O complex O O IP4 O O O O O O O 1-IP7 Kinase Activation dependent IP3 O O Transcription O O O activated Pho80 IP4 O X Pho4 O O Pho85 Kinase activity IP receptor blocked O 3 ENDOPLASMIC Pho81 RETICULUM Ca2+ CYTOPLASM NUCLEUS NUCLEUS mRNA export and translation Insulin secretion and AKT Embryonic development Translation termination Effects of IP kinase deficiency O IPMK (IPK2): Multiple defects, death by embryonic day 10 (mice) O O Insulin IPK1: Cillia are shortened and immotile IP6 AKT resistance causing patterning defects (zebrash) O O Multiple defects, death by Ribosome O embryonic day 8.5 (mice) GleI eRF1 Insulin GSK3β Dbp5 ITPK1 (IP56K): Neural tube -

New Nucleotide Sequence Data on the EMBL File Server
.=) 1991 Oxford University Press Nucleic Acids Research, Vol. 19, No. 2 413 New nucleotide sequence data on the EMBL File Server October 30, 1990 to November 13, 1990 New nucleotide sequence data, available from the EMBL File Server, (see Stoehr, P.J. and Omond, R.A. (1989) Nucleic Acids Res., 17 (16), 6763 -6764), are reported below. Availability of all the newest sequence data is the result of collaboration between.the EMBL Data Library and GenBank' , and data are supplied regularly by both groups. Updates to existing data are not reported here. This report has been prepared by the EMBL Data Library. PRIMATES: Human casein kinase II alpha subunit mRNA, complete cds. Human specific HS5 DNA Lozeman F.J., Litchfield D.W., Piening C., Takio K., Walsh Ueda S., Washio K., Kurosaki K.; K.A., Krebs E.G.; Genomics 8:7-12(1990). X17579 Biochemistry 29:8436-8447(1990). M55268 Human mRNA for heat shock protein HSP27 Human mRNA for actin-binding protein (ABP-280) Carper S.W., Rocheleau T.A., Storm F.K.; Gorlin J.B., Yamin R., Egan S., Stewart M., Stossel T.P., Nucleic Acids Res. 18:6457-6457(1990). X54079 Kwiatkowski D.J., Hartwig J.H.; J. Cell Biol. 111:1089-1105(1990). X53416 Human Ig germline H-chain D-region Dxpl and Dxp'1 genes, 3' Human amiloride-binding protein, complete cds. end. Barbry P., Champe M., Chassande O., Munemitsu S., Champigny Huang C., Stollar B.David.; G., Lingueglia E., Maes P., Frelin C., Tartar A., Ullrich A., Unpublished. M37485 Lazdunski M.; Proc. Natl. Acad. -

Supplementary Materials
Supplementary Materials Figure S1. Differentially abundant spots between the mid-log phase cells grown on xylan or xylose. Red and blue circles denote spots with increased and decreased abundance respectively in the xylan growth condition. The identities of the circled spots are summarized in Table 3. Figure S2. Differentially abundant spots between the stationary phase cells grown on xylan or xylose. Red and blue circles denote spots with increased and decreased abundance respectively in the xylan growth condition. The identities of the circled spots are summarized in Table 4. S2 Table S1. Summary of the non-polysaccharide degrading proteins identified in the B. proteoclasticus cytosol by 2DE/MALDI-TOF. Protein Locus Location Score pI kDa Pep. Cov. Amino Acid Biosynthesis Acetylornithine aminotransferase, ArgD Bpr_I1809 C 1.7 × 10−4 5.1 43.9 11 34% Aspartate/tyrosine/aromatic aminotransferase Bpr_I2631 C 3.0 × 10−14 4.7 43.8 15 46% Aspartate-semialdehyde dehydrogenase, Asd Bpr_I1664 C 7.6 × 10−18 5.5 40.1 17 50% Branched-chain amino acid aminotransferase, IlvE Bpr_I1650 C 2.4 × 10−12 5.2 39.2 13 32% Cysteine synthase, CysK Bpr_I1089 C 1.9 × 10−13 5.0 32.3 18 72% Diaminopimelate dehydrogenase Bpr_I0298 C 9.6 × 10−16 5.6 35.8 16 49% Dihydrodipicolinate reductase, DapB Bpr_I2453 C 2.7 × 10−6 4.9 27.0 9 46% Glu/Leu/Phe/Val dehydrogenase Bpr_I2129 C 1.2 × 10−30 5.4 48.6 31 64% Imidazole glycerol phosphate synthase Bpr_I1240 C 8.0 × 10−3 4.7 22.5 8 44% glutamine amidotransferase subunit Ketol-acid reductoisomerase, IlvC Bpr_I1657 C 3.8 × 10−16 -

Prevalence of Pyruvate Kinase Deficiency
P3513 Prevalence of Red Cell Pyruvate Kinase Deficiency: A Systematic Literature Review Mike Storm1, Matthew H Secrest2*, Courtney Carrington2, Deb Casso2, Keely Gilroy1, Leanne Pladson1, Audra N Boscoe1 1Agios Pharmaceuticals Inc., Cambridge, MA, USA; 2IQVIA Epidemiology & Drug Safety, Seattle, WA and Cambridge, MA, USA; *Affiliation at the time research was conducted BACKGROUND METHODS (continued) RESULTS (continued) RESULTS (continued) • Pyruvate Kinase (PK) deficiency is a rare congenital hemolytic anemia Exclusion Criteria The remaining 34 studies were grouped based on methods and study Among these 4 studies, an important distinction was made between studies characterized by diminished activity of the PK enzyme in red blood population (Table 1). reporting diagnosed prevalence (n=3) and overall disease prevalence cells (RBC).1 • Non-human studies; (diagnosed and undiagnosed PK deficiency; n=1). Table 1. Distribution of extracted studies by type of study (n=34) • Low PK enzyme activity can lead to lifelong chronic hemolysis with • Publications that were not the primary report of the data • Two studies estimated diagnosed PK deficiency prevalence as 3.2 per associated symptoms and complications such as anemia, jaundice, (e.g., literature reviews); Type of study Number million4 and 8.5 per million5 by identifying diagnosed PK deficiency cases of studies gallstones, splenectomy and associated thrombosis, iron overload, and • Studies of PK deficiency prevalence/incidence conducted within a source from source populations of known size. liver cirrhosis.2 Population-based prevalence 2 population of patients with symptoms of PK deficiency such as anemia • We estimated the prevalence of diagnosed PK deficiency in a general • PK deficiency is caused by compound heterozygosity or homozygosity for or jaundice; Molecular PKLR screening in a general population 5 population to be 6.5 per million6 using data from another high-quality study 3 one or more of the >300 known mutations to the PKLR gene. -

Table S1. List of Oligonucleotide Primers Used
Table S1. List of oligonucleotide primers used. Cla4 LF-5' GTAGGATCCGCTCTGTCAAGCCTCCGACC M629Arev CCTCCCTCCATGTACTCcgcGATGACCCAgAGCTCGTTG M629Afwd CAACGAGCTcTGGGTCATCgcgGAGTACATGGAGGGAGG LF-3' GTAGGCCATCTAGGCCGCAATCTCGTCAAGTAAAGTCG RF-5' GTAGGCCTGAGTGGCCCGAGATTGCAACGTGTAACC RF-3' GTAGGATCCCGTACGCTGCGATCGCTTGC Ukc1 LF-5' GCAATATTATGTCTACTTTGAGCG M398Arev CCGCCGGGCAAgAAtTCcgcGAGAAGGTACAGATACGc M398Afwd gCGTATCTGTACCTTCTCgcgGAaTTcTTGCCCGGCGG LF-3' GAGGCCATCTAGGCCATTTACGATGGCAGACAAAGG RF-5' GTGGCCTGAGTGGCCATTGGTTTGGGCGAATGGC RF-3' GCAATATTCGTACGTCAACAGCGCG Nrc2 LF-5' GCAATATTTCGAAAAGGGTCGTTCC M454Grev GCCACCCATGCAGTAcTCgccGCAGAGGTAGAGGTAATC M454Gfwd GATTACCTCTACCTCTGCggcGAgTACTGCATGGGTGGC LF-3' GAGGCCATCTAGGCCGACGAGTGAAGCTTTCGAGCG RF-5' GAGGCCTGAGTGGCCTAAGCATCTTGGCTTCTGC RF-3' GCAATATTCGGTCAACGCTTTTCAGATACC Ipl1 LF-5' GTCAATATTCTACTTTGTGAAGACGCTGC M629Arev GCTCCCCACGACCAGCgAATTCGATagcGAGGAAGACTCGGCCCTCATC M629Afwd GATGAGGGCCGAGTCTTCCTCgctATCGAATTcGCTGGTCGTGGGGAGC LF-3' TGAGGCCATCTAGGCCGGTGCCTTAGATTCCGTATAGC RF-5' CATGGCCTGAGTGGCCGATTCTTCTTCTGTCATCGAC RF-3' GACAATATTGCTGACCTTGTCTACTTGG Ire1 LF-5' GCAATATTAAAGCACAACTCAACGC D1014Arev CCGTAGCCAAGCACCTCGgCCGAtATcGTGAGCGAAG D1014Afwd CTTCGCTCACgATaTCGGcCGAGGTGCTTGGCTACGG LF-3' GAGGCCATCTAGGCCAACTGGGCAAAGGAGATGGA RF-5' GAGGCCTGAGTGGCCGTGCGCCTGTGTATCTCTTTG RF-3' GCAATATTGGCCATCTGAGGGCTGAC Kin28 LF-5' GACAATATTCATCTTTCACCCTTCCAAAG L94Arev TGATGAGTGCTTCTAGATTGGTGTCggcGAAcTCgAGCACCAGGTTG L94Afwd CAACCTGGTGCTcGAgTTCgccGACACCAATCTAGAAGCACTCATCA LF-3' TGAGGCCATCTAGGCCCACAGAGATCCGCTTTAATGC RF-5' CATGGCCTGAGTGGCCAGGGCTAGTACGACCTCG -

Pyruvate Kinase, Rbc
Lab Dept: Chemistry Test Name: PYRUVATE KINASE, RBC General Information Lab Order Codes: PYKI Synonyms: Pyruvate Kinase, Erythrocytes CPT Codes: 84220 – Pyruvate kinase Test Includes: Pyruvate kinase RBC level reported in U/g Hb. Logistics Test Indications: Useful for the workup of cases of nonspherocytic hemolytic anemia, for a family workup to determine inheritance pattern (pyruvate kinase deficiency is autosomal recessive), and for genetic counseling. Lab Testing Sections: Chemistry - Sendouts Referred to: Mayo Medical Laboratories (MML Test: PK) Phone Numbers: MIN Lab: 612-813-6280 STP Lab: 651-220-6550 Test Availability: Daily, 24 hours Turnaround Time: 1 - 4 days, test set up Monday - Saturday Special Instructions: N/A Specimen Specimen Type: Whole blood Container: Yellow top (ACD- Solution B) tube Alternate tube: Lavender (EDTA) top tube Draw Volume: 6 mL (Minimum: 1 mL) blood Processed Volume: Same as Draw Volume Collection: Routine blood collection Special Processing: Lab Staff: Do Not centrifuge. Send whole blood refrigerated in original collection container. Do Not transfer blood to other containers. Store and ship at refrigerated temperatures. Forward promptly. Patient Preparation: None Sample Rejection: Specimen cannot be frozen; mislabeled or unlabeled specimens; gross hemolysis Interpretive Reference Range: ≥12 months: 6.7 – 14.3 U/g Hb Reference values have not been established for patients <12 months of age. Interpretation: Most hemolytic anemias due to pyruvate kinase (PK) deficiency are associated with activity levels less than 40% of mean normal. However, some patients with clinically significant hemolysis can have normal or only mildly decreased PK enzyme activity, which paradoxically may occur in individuals with most severe symptoms. -

Taming the Wild Rubisco: Explorations in Functional Metagenomics
Taming the Wild RubisCO: Explorations in Functional Metagenomics DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Brian Hurin Witte, M.S. Graduate Program in Microbiology The Ohio State University 2012 Dissertation Committee : F. Robert Tabita, Advisor Joseph Krzycki Birgit E. Alber Paul Fuerst Copyright by Brian Hurin Witte 2012 Abstract Ribulose bisphosphate carboxylase/oxygenase (E.C. 4.1.1.39) (RubisCO) is the most abundant protein on Earth and the mechanism by which the vast majority of carbon enters the planet’s biosphere. Despite decades of study, many significant questions about this enzyme remain unanswered. As anthropogenic CO2 levels continue to rise, understanding this key component of the carbon cycle is crucial to forecasting feedback circuits, as well as to engineering food and fuel crops to produce more biomass with few inputs of increasingly scarce resources. This study demonstrates three means of investigating the natural diversity of RubisCO. Chapter 1 builds on existing DNA sequence-based techniques of gene discovery and shows that RubisCO from uncultured organisms can be used to complement growth in a RubisCO-deletion strain of autotrophic bacteria. In a few short steps, the time-consuming work of bringing an autotrophic organism in to pure culture can be circumvented. Chapter 2 details a means of entirely bypassing the bias inherent in sequence-based gene discovery by using selection of RubisCO genes from a metagenomic library. Chapter 3 provides a more in-depth study of the RubisCO from the methanogenic archaeon Methanococcoides burtonii. -

Ceramide Kinase Is Upregulated in Metastatic Breast Cancer Cells and Contributes to Migration and Invasion by Activation of PI 3-Kinase and Akt
International Journal of Molecular Sciences Article Ceramide Kinase Is Upregulated in Metastatic Breast Cancer Cells and Contributes to Migration and Invasion by Activation of PI 3-Kinase and Akt 1,2, 1, 2 2 Stephanie Schwalm y, Martin Erhardt y, Isolde Römer , Josef Pfeilschifter , Uwe Zangemeister-Wittke 1,* and Andrea Huwiler 1,* 1 Institute of Pharmacology, University of Bern, Inselspital, INO-F, CH-3010 Bern, Switzerland; [email protected] (S.S.); [email protected] (M.E.) 2 Institute of General Pharmacology and Toxicology, University Hospital Frankfurt am Main, Goethe-University, Theodor-Stern Kai 7, D-60590 Frankfurt am Main, Germany; [email protected] (I.R.); [email protected] (J.P.) * Correspondence: [email protected] (U.Z.-W.); [email protected] (A.H.); Tel.: +41-31-6323214 (A.H.) These authors contributed equally to this work. y Received: 29 November 2019; Accepted: 12 February 2020; Published: 19 February 2020 Abstract: Ceramide kinase (CerK) is a lipid kinase that converts the proapoptotic ceramide to ceramide 1-phosphate, which has been proposed to have pro-malignant properties and regulate cell responses such as proliferation, migration, and inflammation. We used the parental human breast cancer cell line MDA-MB-231 and two single cell progenies derived from lung and bone metastasis upon injection of the parental cells into immuno-deficient mice. The lung and the bone metastatic cell lines showed a marked upregulation of CerK mRNA and activity when compared to the parental cell line. The metastatic cells also had increased migratory and invasive activity, which was dose-dependently reduced by the selective CerK inhibitor NVP-231.