Product Sheet Info
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
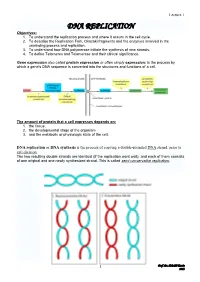
DNA REPLICATION Objectives: 1
Lecture 1 DNA REPLICATION Objectives: 1. To understand the replication process and where it occurs in the cell cycle. 2. To describe the Replication Fork, Okazaki fragments and the enzymes involved in the unwinding process and replication. 3. To understand how DNA polymerase initiate the synthesis of new strands. 4. To define Telomeres and Telomerase and their clinical significance. Gene expression also called protein expression or often simply expression: is the process by which a gene's DNA sequence is converted into the structures and functions of a cell. The amount of protein that a cell expresses depends on: 1. the tissue, 2. the developmental stage of the organism 3. and the metabolic or physiologic state of the cell. DNA replication or DNA synthesis is the process of copying a double-stranded DNA strand, prior to cell division. The two resulting double strands are identical (if the replication went well), and each of them consists of one original and one newly synthesized strand. This is called semi conservative replication. 1 Prof. Dr. H.D.El-Yassin 2013 Lecture 1 The process of replication consists of three steps, initiation, replication and termination. 1. Prokaryotic replication Basic Requirement for DNA Synthesis 1. Substrates: the four deoxy nucleosides triphosphates are needed as substrates for DNA synthesis. Cleavage of the high-energy phosphate bond between the α and β phosphates provides the energy for the addition of the nucleotide. 2. Template: DNA replication cannot occur without a template. A template is required to direct the addition of the appropriate complementary deoxynucleotide to the newly synthesized DNA strand. -
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli
DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Kath, James Evon. 2016. DNA Polymerase Exchange and Lesion Bypass in Escherichia Coli. Doctoral dissertation, Harvard University, Graduate School of Arts & Sciences. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:26718716 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA ! ! ! ! ! ! ! DNA!polymerase!exchange!and!lesion!bypass!in!Escherichia)coli! ! A!dissertation!presented! by! James!Evon!Kath! to! The!Committee!on!Higher!Degrees!in!Biophysics! ! in!partial!fulfillment!of!the!requirements! for!the!degree!of! Doctor!of!Philosophy! in!the!subject!of! Biophysics! ! Harvard!University! Cambridge,!Massachusetts! October!2015! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ©!2015!L!James!E.!Kath.!Some!Rights!Reserved.! ! This!work!is!licensed!under!the!Creative!Commons!Attribution!3.0!United!States!License.!To! view!a!copy!of!this!license,!visit:!http://creativecommons.org/licenses/By/3.0/us! ! ! Dissertation!Advisor:!Professor!Joseph!J.!Loparo! ! ! !!!!!!!!James!Evon!Kath! ! DNA$polymerase$exchange$and$lesion$bypass$in$Escherichia)coli$ $ Abstract$ ! Translesion! synthesis! (TLS)! alleviates! -

Folic Acid Antagonists: Antimicrobial and Immunomodulating Mechanisms and Applications
International Journal of Molecular Sciences Review Folic Acid Antagonists: Antimicrobial and Immunomodulating Mechanisms and Applications Daniel Fernández-Villa 1, Maria Rosa Aguilar 1,2 and Luis Rojo 1,2,* 1 Instituto de Ciencia y Tecnología de Polímeros, Consejo Superior de Investigaciones Científicas, CSIC, 28006 Madrid, Spain; [email protected] (D.F.-V.); [email protected] (M.R.A.) 2 Consorcio Centro de Investigación Biomédica en Red de Bioingeniería, Biomateriales y Nanomedicina, 28029 Madrid, Spain * Correspondence: [email protected]; Tel.: +34-915-622-900 Received: 18 September 2019; Accepted: 7 October 2019; Published: 9 October 2019 Abstract: Bacterial, protozoan and other microbial infections share an accelerated metabolic rate. In order to ensure a proper functioning of cell replication and proteins and nucleic acids synthesis processes, folate metabolism rate is also increased in these cases. For this reason, folic acid antagonists have been used since their discovery to treat different kinds of microbial infections, taking advantage of this metabolic difference when compared with human cells. However, resistances to these compounds have emerged since then and only combined therapies are currently used in clinic. In addition, some of these compounds have been found to have an immunomodulatory behavior that allows clinicians using them as anti-inflammatory or immunosuppressive drugs. Therefore, the aim of this review is to provide an updated state-of-the-art on the use of antifolates as antibacterial and immunomodulating agents in the clinical setting, as well as to present their action mechanisms and currently investigated biomedical applications. Keywords: folic acid antagonists; antifolates; antibiotics; antibacterials; immunomodulation; sulfonamides; antimalarial 1. -

(12) Patent Application Publication (10) Pub. No.: US 2013/0089535 A1 Yamashiro Et Al
US 2013 0089535A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2013/0089535 A1 Yamashiro et al. (43) Pub. Date: Apr. 11, 2013 (54) AGENT FOR REDUCING ACETALDEHYDE Publication Classification NORAL CAVITY (51) Int. Cl. (75) Inventors: Kan Yamashiro, Kakamigahara-shi (JP); A68/66 (2006.01) Takahumi Koyama, Kakamigahara-shi A638/51 (2006.01) (JP) A61O 11/00 (2006.01) A638/44 (2006.01) Assignee: AMANOENZYME INC., Nagoya-shi (52) U.S. Cl. (73) CPC. A61K 8/66 (2013.01); A61K 38/44 (2013.01); (JP) A61 K38/51 (2013.01); A61O II/00 (2013.01) (21) Appl. No.: 13/703,451 USPC .......... 424/94.4; 424/94.5; 435/191: 435/232 (22) PCT Fled: Jun. 7, 2011 (57) ABSTRACT Disclosed herein is a novel enzymatic agent effective in (86) PCT NO.: PCT/UP2011/062991 reducing acetaldehyde in the oral cavity. It has been found S371 (c)(1), that an aldehyde dehydrogenase derived from a microorgan (2), (4) Date: Dec. 11, 2012 ism belonging to the genus Saccharomyces and a threonine aldolase derived from Escherichia coli are effective in reduc (30) Foreign Application Priority Data ing low concentrations of acetaldehyde. Therefore, an agent for reducing acetaldehyde in the oral cavity is provided, Jun. 19, 2010 (JP) ................................. 2010-140O26 which contains these enzymes as active ingredients. Patent Application Publication Apr. 11, 2013 Sheet 1 of 2 US 2013/0089535 A1 FIG 1) 10.5 1 0 9.9.5 8. 5 CONTROL TA AD (BSA) ENZYME Patent Application Publication Apr. 11, 2013 Sheet 2 of 2 US 2013/0089535 A1 FIG 2) 110 the CONTROL (BSA) 100 354. -

Enzymatic Encoding Methods for Efficient Synthesis Of
(19) TZZ__T (11) EP 1 957 644 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C12N 15/10 (2006.01) C12Q 1/68 (2006.01) 01.12.2010 Bulletin 2010/48 C40B 40/06 (2006.01) C40B 50/06 (2006.01) (21) Application number: 06818144.5 (86) International application number: PCT/DK2006/000685 (22) Date of filing: 01.12.2006 (87) International publication number: WO 2007/062664 (07.06.2007 Gazette 2007/23) (54) ENZYMATIC ENCODING METHODS FOR EFFICIENT SYNTHESIS OF LARGE LIBRARIES ENZYMVERMITTELNDE KODIERUNGSMETHODEN FÜR EINE EFFIZIENTE SYNTHESE VON GROSSEN BIBLIOTHEKEN PROCEDES DE CODAGE ENZYMATIQUE DESTINES A LA SYNTHESE EFFICACE DE BIBLIOTHEQUES IMPORTANTES (84) Designated Contracting States: • GOLDBECH, Anne AT BE BG CH CY CZ DE DK EE ES FI FR GB GR DK-2200 Copenhagen N (DK) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • DE LEON, Daen SK TR DK-2300 Copenhagen S (DK) Designated Extension States: • KALDOR, Ditte Kievsmose AL BA HR MK RS DK-2880 Bagsvaerd (DK) • SLØK, Frank Abilgaard (30) Priority: 01.12.2005 DK 200501704 DK-3450 Allerød (DK) 02.12.2005 US 741490 P • HUSEMOEN, Birgitte Nystrup DK-2500 Valby (DK) (43) Date of publication of application: • DOLBERG, Johannes 20.08.2008 Bulletin 2008/34 DK-1674 Copenhagen V (DK) • JENSEN, Kim Birkebæk (73) Proprietor: Nuevolution A/S DK-2610 Rødovre (DK) 2100 Copenhagen 0 (DK) • PETERSEN, Lene DK-2100 Copenhagen Ø (DK) (72) Inventors: • NØRREGAARD-MADSEN, Mads • FRANCH, Thomas DK-3460 Birkerød (DK) DK-3070 Snekkersten (DK) • GODSKESEN, -

On the Active Site Thiol of Y-Glutamylcysteine Synthetase
Proc. Natl. Acad. Sci. USA Vol. 85, pp. 2464-2468, April 1988 Biochemistry On the active site thiol of y-glutamylcysteine synthetase: Relationships to catalysis, inhibition, and regulation (glutathione/cystamine/Escherichia coli/kidney/enzyme inactivation) CHIN-SHIou HUANG, WILLIAM R. MOORE, AND ALTON MEISTER Cornell University Medical College, Department of Biochemistry, 1300 York Avenue, New York, NY 10021 Contributed by Alton Meister, December 4, 1987 ABSTRACT y-Glutamylcysteine synthetase (glutamate- dithiothreitol, suggesting that cystamine forms a mixed cysteine ligase; EC 6.3.2.2) was isolated from an Escherichia disulfide between cysteamine and an enzyme thiol (15). coli strain enriched in the gene for this enzyme by recombinant Inactivation of the enzyme by the L- and D-isomers of DNA techniques. The purified enzyme has a specific activity of 3-amino-1-chloro-2-pentanone, as well as that by cystamine, 1860 units/mg and a molecular weight of 56,000. Comparison is prevented by L-glutamate (14). Treatment of the enzyme of the E. coli enzyme with the well-characterized rat kidney with cystamine prevents its interaction with the sulfoxi- enzyme showed that these enzymes have similar catalytic prop- mines. Titration of the enzyme with 5,5'-dithiobis(2- erties (apparent Km values, substrate specificities, turnover nitrobenzoate) reveals that the enzyme has a single exposed numbers). Both enzymes are feedback-inhibited by glutathione thiol that reacts with this reagent without affecting activity but not by y-glutamyl-a-aminobutyrylglycine; the data indicate (16). 5,5'-Dithiobis(2-nitrobenzoate) does not interact with that glutathione binds not only at the glutamate binding site but the thiol that reacts with cystamine. -

Control Engineering Perspective on Genome-Scale Metabolic Modeling
Control Engineering Perspective on Genome-Scale Metabolic Modeling by Andrew Louis Damiani A dissertation submitted to the Graduate Faculty of Auburn University in partial fulfillment of the requirements for the Degree of Doctor of Philosophy Auburn, Alabama December 12, 2015 Key words: Scheffersomyces stipitis, Flux Balance Analysis, Genome-scale metabolic models, System Identification Framework, Model Validation, Phenotype Phase Plane Analysis Copyright 2015 by Andrew Damiani Approved by Jin Wang, Chair, Associate Professor of Chemical Engineering Q. Peter He, Associate Professor of Chemical Engineering, Tuskegee University Thomas W. Jeffries, Professor of Bacteriology, Emeritus; University of Wisconsin-Madison Allan E. David, Assistant Professor of Chemical Engineering Yoon Y. Lee, Professor of Chemical Engineering Abstract Fossil fuels impart major problems on the global economy and have detrimental effects to the environment, which has caused a world-wide initiative of producing renewable fuels. Lignocellulosic bioethanol for renewable energy has recently gained attention, because it can overcome the limitations that first generation biofuels impose. Nonetheless, in order to have this process commercialized, the biological conversion of pentose sugars, mainly xylose, needs to be improved. Scheffersomyces stipitis has a physiology that makes it a valuable candidate for lignocellulosic bioethanol production, and lately has provided genes for designing recombinant Saccharomyces cerevisiae. In this study, a system biology approach was taken to understand the relationship of the genotype to phenotype, whereby genome-scale metabolic models (GSMMs) are used in conjunction with constraint-based modeling. The major restriction of GSMMs is having an accurate methodology for validation and evaluation. This is due to the size and complexity of the models. -

Protein Expression Profile of Gluconacetobacter Diazotrophicus PAL5, a Sugarcane Endophytic Plant Growth-Promoting Bacterium
Proteomics 2008, 8, 1631–1644 DOI 10.1002/pmic.200700912 1631 RESEARCH ARTICLE Protein expression profile of Gluconacetobacter diazotrophicus PAL5, a sugarcane endophytic plant growth-promoting bacterium Leticia M. S. Lery1, 2, Ana Coelho1, 3, Wanda M. A. von Kruger1, 2, Mayla S. M. Gonc¸alves1, 3, Marise F. Santos1, 4, Richard H. Valente1, 5, Eidy O. Santos1, 3, Surza L. G. Rocha1, 5, Jonas Perales1, 5, Gilberto B. Domont1, 4, Katia R. S. Teixeira1, 6 and Paulo M. Bisch1, 2 1 Rio de Janeiro Proteomics Network, Rio de Janeiro, Brazil 2 Unidade Multidisciplinar de Genômica, Instituto de Biofísica Carlos Chagas Filho, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil 3 Departamento de Genética, Instituto de Biologia, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil 4 Laboratório de Química de Proteínas, Departamento de Bioquímica, Instituto de Química, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil 5 Laboratório de Toxinologia, Departamento de Fisiologia e Farmacodinâmica- Instituto Oswaldo Cruz- Fundac¸ão Oswaldo Cruz, Rio de Janeiro, Rio de Janeiro, Brazil 6 Laboratório de Genética e Bioquímica, Embrapa Agrobiologia, Seropédica, Brazil This is the first broad proteomic description of Gluconacetobacter diazotrophicus, an endophytic Received: September 25, 2007 bacterium, responsible for the major fraction of the atmospheric nitrogen fixed in sugarcane in Revised: December 18, 2007 tropical regions. Proteomic coverage of G. diazotrophicus PAL5 was obtained by two independent Accepted: December 19, 2007 approaches: 2-DE followed by MALDI-TOF or TOF-TOF MS and 1-DE followed by chromatog- raphy in a C18 column online coupled to an ESI-Q-TOF or ESI-IT mass spectrometer. -

The Phylogenetic Extent of Metabolic Enzymes and Pathways José Manuel Peregrin-Alvarez, Sophia Tsoka, Christos A
Downloaded from genome.cshlp.org on October 8, 2021 - Published by Cold Spring Harbor Laboratory Press Letter The Phylogenetic Extent of Metabolic Enzymes and Pathways José Manuel Peregrin-Alvarez, Sophia Tsoka, Christos A. Ouzounis1 Computational Genomics Group, The European Bioinformatics Institute, EMBL Cambridge Outstation, Cambridge CB10 1SD, UK The evolution of metabolic enzymes and pathways has been a subject of intense study for more than half a century. Yet, so far, previous studies have focused on a small number of enzyme families or biochemical pathways. Here, we examine the phylogenetic distribution of the full-known metabolic complement of Escherichia coli, using sequence comparison against taxa-specific databases. Half of the metabolic enzymes have homologs in all domains of life, representing families involved in some of the most fundamental cellular processes. We thus show for the first time and in a comprehensive way that metabolism is conserved at the enzyme level. In addition, our analysis suggests that despite the sequence conservation and the extensive phylogenetic distribution of metabolic enzymes, their groupings into biochemical pathways are much more variable than previously thought. One of the fundamental tenets in molecular biology was ex- reliable source of metabolic information. The EcoCyc data- pressed by Monod, in his famous phrase “What is true for base holds information about the full genome and all known Escherichia coli is true for the elephant” (Jacob 1988). For a metabolic pathways of Escherichia coli (Karp et al. 2000). Re- long time, this statement has inspired generations of molecu- cently, the database has been used to represent computational lar biologists, who have used Bacteria as model organisms to predictions of other organisms (Karp 2001). -

Polymerase Ribozyme with Promoter Recognition
In vitro Evolution of a Processive Clamping RNA Polymerase Ribozyme with Promoter Recognition by Razvan Cojocaru BSc, Simon Fraser University, 2014 Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in the Department of Molecular Biology and Biochemistry Faculty of Science © Razvan Cojocaru 2021 SIMON FRASER UNIVERSITY Summer 2021 Copyright in this work is held by the author. Please ensure that any reproduction or re-use is done in accordance with the relevant national copyright legislation. Declaration of Committee Name: Razvan Cojocaru Degree: Doctor of Philosophy Title: In vitro Evolution of a Processive Clamping RNA Polymerase Ribozyme with Promoter Recognition Committee: Chair: Lisa Craig Professor, Molecular Biology and Biochemistry Peter Unrau Supervisor Professor, Molecular Biology and Biochemistry Dipankar Sen Committee Member Professor, Molecular Biology and Biochemistry Michel Leroux Committee Member Professor, Molecular Biology and Biochemistry Mani Larijani Internal Examiner Associate Professor, Molecular Biology and Biochemistry Gerald Joyce External Examiner Professor, Jack H. Skirball Center for Chemical Biology and Proteomics Salk Institute for Biological Studies Date Defended/Approved: August 12, 2021 ii Abstract The RNA World hypothesis proposes that the early evolution of life began with RNAs that can serve both as carriers of genetic information and as catalysts. Later in evolution, these functions were gradually replaced by DNA and enzymatic proteins in cellular biology. I start by reviewing the naturally occurring catalytic RNAs, ribozymes, as they play many important roles in biology today. These ribozymes are central to protein synthesis and the regulation of gene expression, creating a landscape that strongly supports an early RNA World. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -

K319-100 Gluconokinase Activity Assay Kit (Colorimetric)
FOR RESEARCH USE ONLY! Gluconokinase Activity Assay Kit (Colorimetric) 7/16 (Catalog # K319-100; 100 assays; Store at -20°C) I. Introduction: Gluconokinase (ATP:D-gluconate 6-phosphotransferase or Gluconate Kinase; EC:2.7.1.12) is a key enzyme for Gluconate degradation pathway. In E. coli and yeast, Gluconokinase can convert gluconate into 6-Phosphate-D-Gluconate in an ATP dependent manner. Through Hexose Monophosphate Shunt (HMS) pathway, 6-Phosphate-D-Gluconate generates ribose-6-phosphate, which is critical for nucleotides and nucleic acid synthesis. Little is known of the mechanism of gluconate metabolism in humans despite its widespread use in medicine and consumer products. BioVision’s Gluconokinase Assay kit provides a quick and easy way for monitoring Gluconokinase activity in a variety of samples. In this kit, Gluconokinase converts Gluconate into 6-Phosphate-D-Gluconate in an ATP dependent manner. 6-Phosphate-D-Gluconate and ADP in turn undergoe a series of reactions to form an intermediate, which reacts with the probe to form a colored product with strong absorbance (OD 450 nm). The assay is simple, sensitive, and high-throughput adaptable. Detection limit: < 0.1mU. Gluconokinase D-Gluconate + ATP 6-Phosphate-D-Gluconate + ADP Intermediate + Probe Color Product (OD 450 nm) II. Application: Measurement of Gluconokinase activity in various samples Mechanistic study of Pentose Phosphate Pathway III. Sample Type: Prokaryote such as: E.coli Animal tissues such as liver, kidney, etc. Adherent or suspension cells. IV. Kit Contents: Components K319-100 Cap Code Part Number Gluconokinase Assay Buffer 25 ml WM K319-100-1 Gluconokinase Substrate 1 Vial Blue K319-100-2 ATP 1 Vial Orange K319-100-3 Gluconokinase Converting Enzyme 1 Vial Purple K319-100-4 Gluconokinase Developer 1 Vial Green K319-100-5 Gluconokinase Probe 1 Vial Red K319-100-6 NADH Standard 1 Vial Yellow K319-100-7 Gluconokinase Positive Control 1 Vial Brown K319-100-8 V.