Glucokinase September 15, 2005
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Isozymes of Pyruvate Kinase in Liver and Hepatomas of the Rat1
[CANCER RESEARCH 34, 1439-1446, June 1974] Isozymes of Pyruvate Kinase in Liver and Hepatomas of the Rat1 Francis A. Farina,2 Jennie B. Shatton, Harold P. Morris, and Sidney Weinhouse The Fels Research Institute and the Department of Biochemistry, Temple University School oj Medicine, Philadelphia. Pennsylvania IV140 (F. A. F., J. B. S.. S. W.\, and the Department of Biochemistry. Howard University School of Medicine. Washington. D. C. 20001 [H. P. M .\ SUMMARY 23, 32, 41, 57). These alterations involve the replacement of those isozymes that are under dietary and hormonal control Pyruvate kinase (PK) (EC 2.7.1.40) isozymes were assayed by the host, and that have important metabolic functions in in normal rat liver and a series of transplantable rat the adult differentiated liver by other isozymes which are hepatomas ranging widely in growth rate and degree of normally either low in, or absent from, the adult tissue. As differentiation, with the use of gradient elution by chloride part of an ongoing study of this phenomenon, we have ion from columns of DEAE-cellulose. In agreement with examined the alteration of PK3 (EC 2.7.1.40) isozymes in other studies, three noninterconvertible forms were found in the Morris hepatomas (30. 31), a series of chemically rat tissues: isozyme I, the major form in adult rat liver: induced, transplantable rat hepatomas ranging widely in isozyme II, the sole form in heart and skeletal muscle: and growth rate and degree of differentiation. This enzyme isozyme III, the sole form in poorly differentiated hepato occupies a key position in the metabolism of cells and, as we mas, the major form in normal kidney and lung, and the pointed out previously (27, 28. -

Pyruvate Kinase: Function, Regulation and Role in Cancer
Pyruvate kinase: Function, regulation and role in cancer The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Israelsen, William J., and Matthew G. Vander Heiden. “Pyruvate Kinase: Function, Regulation and Role in Cancer.” Seminars in Cell & Developmental Biology 43 (2015): 43–51. As Published http://dx.doi.org/10.1016/j.semcdb.2015.08.004 Publisher Elsevier Version Author's final manuscript Citable link http://hdl.handle.net/1721.1/105833 Terms of Use Creative Commons Attribution-NonCommercial-NoDerivs License Detailed Terms http://creativecommons.org/licenses/by-nc-nd/4.0/ HHS Public Access Author manuscript Author Manuscript Author ManuscriptSemin Cell Author Manuscript Dev Biol. Author Author Manuscript manuscript; available in PMC 2016 August 13. Published in final edited form as: Semin Cell Dev Biol. 2015 July ; 43: 43–51. doi:10.1016/j.semcdb.2015.08.004. Pyruvate kinase: function, regulation and role in cancer William J. Israelsena,1,* and Matthew G. Vander Heidena,b,* aKoch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, Cambridge, MA 02139, USA bDepartment of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02115, USA Abstract Pyruvate kinase is an enzyme that catalyzes the conversion of phosphoenolpyruvate and ADP to pyruvate and ATP in glycolysis and plays a role in regulating cell metabolism. There are four mammalian pyruvate kinase isoforms with unique tissue expression patterns and regulatory properties. The M2 isoform of pyruvate kinase (PKM2) supports anabolic metabolism and is expressed both in cancer and normal tissue. The enzymatic activity of PKM2 is allosterically regulated by both intracellular signaling pathways and metabolites; PKM2 thus integrates signaling and metabolic inputs to modulate glucose metabolism according to the needs of the cell. -
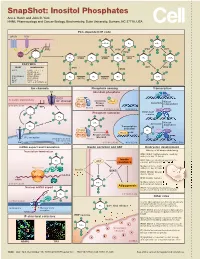
Snapshot: Inositol Phosphates Ace J
SnapShot: Inositol Phosphates Ace J. Hatch and John D. York HHMI, Pharmacology and Cancer Biology, Biochemistry, Duke University, Durham, NC 27710, USA PLC-dependent IP code GPCR RTK O O O O O O 5-PP-IP4 IP4 5-IP7 O O O O O O PIP2 O IP6K O IP6K O VIP1 O O O 2 O ITPK1 O 13 O PLC 2 O O O O O O O O 4 6 13 O 5 IP3 IPMK IP4 IPMK IP5 IPK1 IP6 1,5-IP8 4 6 O O 5 O O O O O O O O ENZYMES O O O O O O YEAST MAMMALIAN IP3K VIP1 IP6K IPMK PLC1 PLCβ, γ, δ, ε, ζ, η O - IP3KA, B, C - ITPK1 (IP56K) O O O O O O O O IPK2(ARG82) IPMK (IPK2) IP4 IP3 IP4 1-IP7 IPK1 IPK1 (IP5K) INPP5 ITPK1 KCS1 IP6K1, 2, 3 O O O O O O VIP1 VIP1, 2 (PPIP5K1, 2) O O Ion channels Phosphate sensing Transcription Cl- Abundant phosphate MCM1 ARG80 CIC3 P PLASMA MEMBRANE - Pho80 Cl channel Pho4 Kinase Kinase Assembly Pho85 independent CYTOPLASM activity 2 O PIP2 Pho81 13 CYTOPLASM NUCLEUS IPK2 ARG81 4 6 Phosphate starvation MCM1-ArgR O 5 O complex O O IP4 O O O O O O O 1-IP7 Kinase Activation dependent IP3 O O Transcription O O O activated Pho80 IP4 O X Pho4 O O Pho85 Kinase activity IP receptor blocked O 3 ENDOPLASMIC Pho81 RETICULUM Ca2+ CYTOPLASM NUCLEUS NUCLEUS mRNA export and translation Insulin secretion and AKT Embryonic development Translation termination Effects of IP kinase deficiency O IPMK (IPK2): Multiple defects, death by embryonic day 10 (mice) O O Insulin IPK1: Cillia are shortened and immotile IP6 AKT resistance causing patterning defects (zebrash) O O Multiple defects, death by Ribosome O embryonic day 8.5 (mice) GleI eRF1 Insulin GSK3β Dbp5 ITPK1 (IP56K): Neural tube -

Prevalence of Pyruvate Kinase Deficiency
P3513 Prevalence of Red Cell Pyruvate Kinase Deficiency: A Systematic Literature Review Mike Storm1, Matthew H Secrest2*, Courtney Carrington2, Deb Casso2, Keely Gilroy1, Leanne Pladson1, Audra N Boscoe1 1Agios Pharmaceuticals Inc., Cambridge, MA, USA; 2IQVIA Epidemiology & Drug Safety, Seattle, WA and Cambridge, MA, USA; *Affiliation at the time research was conducted BACKGROUND METHODS (continued) RESULTS (continued) RESULTS (continued) • Pyruvate Kinase (PK) deficiency is a rare congenital hemolytic anemia Exclusion Criteria The remaining 34 studies were grouped based on methods and study Among these 4 studies, an important distinction was made between studies characterized by diminished activity of the PK enzyme in red blood population (Table 1). reporting diagnosed prevalence (n=3) and overall disease prevalence cells (RBC).1 • Non-human studies; (diagnosed and undiagnosed PK deficiency; n=1). Table 1. Distribution of extracted studies by type of study (n=34) • Low PK enzyme activity can lead to lifelong chronic hemolysis with • Publications that were not the primary report of the data • Two studies estimated diagnosed PK deficiency prevalence as 3.2 per associated symptoms and complications such as anemia, jaundice, (e.g., literature reviews); Type of study Number million4 and 8.5 per million5 by identifying diagnosed PK deficiency cases of studies gallstones, splenectomy and associated thrombosis, iron overload, and • Studies of PK deficiency prevalence/incidence conducted within a source from source populations of known size. liver cirrhosis.2 Population-based prevalence 2 population of patients with symptoms of PK deficiency such as anemia • We estimated the prevalence of diagnosed PK deficiency in a general • PK deficiency is caused by compound heterozygosity or homozygosity for or jaundice; Molecular PKLR screening in a general population 5 population to be 6.5 per million6 using data from another high-quality study 3 one or more of the >300 known mutations to the PKLR gene. -

Table S1. List of Oligonucleotide Primers Used
Table S1. List of oligonucleotide primers used. Cla4 LF-5' GTAGGATCCGCTCTGTCAAGCCTCCGACC M629Arev CCTCCCTCCATGTACTCcgcGATGACCCAgAGCTCGTTG M629Afwd CAACGAGCTcTGGGTCATCgcgGAGTACATGGAGGGAGG LF-3' GTAGGCCATCTAGGCCGCAATCTCGTCAAGTAAAGTCG RF-5' GTAGGCCTGAGTGGCCCGAGATTGCAACGTGTAACC RF-3' GTAGGATCCCGTACGCTGCGATCGCTTGC Ukc1 LF-5' GCAATATTATGTCTACTTTGAGCG M398Arev CCGCCGGGCAAgAAtTCcgcGAGAAGGTACAGATACGc M398Afwd gCGTATCTGTACCTTCTCgcgGAaTTcTTGCCCGGCGG LF-3' GAGGCCATCTAGGCCATTTACGATGGCAGACAAAGG RF-5' GTGGCCTGAGTGGCCATTGGTTTGGGCGAATGGC RF-3' GCAATATTCGTACGTCAACAGCGCG Nrc2 LF-5' GCAATATTTCGAAAAGGGTCGTTCC M454Grev GCCACCCATGCAGTAcTCgccGCAGAGGTAGAGGTAATC M454Gfwd GATTACCTCTACCTCTGCggcGAgTACTGCATGGGTGGC LF-3' GAGGCCATCTAGGCCGACGAGTGAAGCTTTCGAGCG RF-5' GAGGCCTGAGTGGCCTAAGCATCTTGGCTTCTGC RF-3' GCAATATTCGGTCAACGCTTTTCAGATACC Ipl1 LF-5' GTCAATATTCTACTTTGTGAAGACGCTGC M629Arev GCTCCCCACGACCAGCgAATTCGATagcGAGGAAGACTCGGCCCTCATC M629Afwd GATGAGGGCCGAGTCTTCCTCgctATCGAATTcGCTGGTCGTGGGGAGC LF-3' TGAGGCCATCTAGGCCGGTGCCTTAGATTCCGTATAGC RF-5' CATGGCCTGAGTGGCCGATTCTTCTTCTGTCATCGAC RF-3' GACAATATTGCTGACCTTGTCTACTTGG Ire1 LF-5' GCAATATTAAAGCACAACTCAACGC D1014Arev CCGTAGCCAAGCACCTCGgCCGAtATcGTGAGCGAAG D1014Afwd CTTCGCTCACgATaTCGGcCGAGGTGCTTGGCTACGG LF-3' GAGGCCATCTAGGCCAACTGGGCAAAGGAGATGGA RF-5' GAGGCCTGAGTGGCCGTGCGCCTGTGTATCTCTTTG RF-3' GCAATATTGGCCATCTGAGGGCTGAC Kin28 LF-5' GACAATATTCATCTTTCACCCTTCCAAAG L94Arev TGATGAGTGCTTCTAGATTGGTGTCggcGAAcTCgAGCACCAGGTTG L94Afwd CAACCTGGTGCTcGAgTTCgccGACACCAATCTAGAAGCACTCATCA LF-3' TGAGGCCATCTAGGCCCACAGAGATCCGCTTTAATGC RF-5' CATGGCCTGAGTGGCCAGGGCTAGTACGACCTCG -

Pyruvate Kinase, Rbc
Lab Dept: Chemistry Test Name: PYRUVATE KINASE, RBC General Information Lab Order Codes: PYKI Synonyms: Pyruvate Kinase, Erythrocytes CPT Codes: 84220 – Pyruvate kinase Test Includes: Pyruvate kinase RBC level reported in U/g Hb. Logistics Test Indications: Useful for the workup of cases of nonspherocytic hemolytic anemia, for a family workup to determine inheritance pattern (pyruvate kinase deficiency is autosomal recessive), and for genetic counseling. Lab Testing Sections: Chemistry - Sendouts Referred to: Mayo Medical Laboratories (MML Test: PK) Phone Numbers: MIN Lab: 612-813-6280 STP Lab: 651-220-6550 Test Availability: Daily, 24 hours Turnaround Time: 1 - 4 days, test set up Monday - Saturday Special Instructions: N/A Specimen Specimen Type: Whole blood Container: Yellow top (ACD- Solution B) tube Alternate tube: Lavender (EDTA) top tube Draw Volume: 6 mL (Minimum: 1 mL) blood Processed Volume: Same as Draw Volume Collection: Routine blood collection Special Processing: Lab Staff: Do Not centrifuge. Send whole blood refrigerated in original collection container. Do Not transfer blood to other containers. Store and ship at refrigerated temperatures. Forward promptly. Patient Preparation: None Sample Rejection: Specimen cannot be frozen; mislabeled or unlabeled specimens; gross hemolysis Interpretive Reference Range: ≥12 months: 6.7 – 14.3 U/g Hb Reference values have not been established for patients <12 months of age. Interpretation: Most hemolytic anemias due to pyruvate kinase (PK) deficiency are associated with activity levels less than 40% of mean normal. However, some patients with clinically significant hemolysis can have normal or only mildly decreased PK enzyme activity, which paradoxically may occur in individuals with most severe symptoms. -

Ceramide Kinase Is Upregulated in Metastatic Breast Cancer Cells and Contributes to Migration and Invasion by Activation of PI 3-Kinase and Akt
International Journal of Molecular Sciences Article Ceramide Kinase Is Upregulated in Metastatic Breast Cancer Cells and Contributes to Migration and Invasion by Activation of PI 3-Kinase and Akt 1,2, 1, 2 2 Stephanie Schwalm y, Martin Erhardt y, Isolde Römer , Josef Pfeilschifter , Uwe Zangemeister-Wittke 1,* and Andrea Huwiler 1,* 1 Institute of Pharmacology, University of Bern, Inselspital, INO-F, CH-3010 Bern, Switzerland; [email protected] (S.S.); [email protected] (M.E.) 2 Institute of General Pharmacology and Toxicology, University Hospital Frankfurt am Main, Goethe-University, Theodor-Stern Kai 7, D-60590 Frankfurt am Main, Germany; [email protected] (I.R.); [email protected] (J.P.) * Correspondence: [email protected] (U.Z.-W.); [email protected] (A.H.); Tel.: +41-31-6323214 (A.H.) These authors contributed equally to this work. y Received: 29 November 2019; Accepted: 12 February 2020; Published: 19 February 2020 Abstract: Ceramide kinase (CerK) is a lipid kinase that converts the proapoptotic ceramide to ceramide 1-phosphate, which has been proposed to have pro-malignant properties and regulate cell responses such as proliferation, migration, and inflammation. We used the parental human breast cancer cell line MDA-MB-231 and two single cell progenies derived from lung and bone metastasis upon injection of the parental cells into immuno-deficient mice. The lung and the bone metastatic cell lines showed a marked upregulation of CerK mRNA and activity when compared to the parental cell line. The metastatic cells also had increased migratory and invasive activity, which was dose-dependently reduced by the selective CerK inhibitor NVP-231. -

Inhibition of Spinach Phosphoribulokinase by DL-Glyceraldehyde by ANTONI R
Biochem. J. (1976) 153, 613-619 613 Printed in Great Britain Inhibition of Spinach Phosphoribulokinase by DL-Glyceraldehyde By ANTONI R. SLABAS and DAVID A. WALKER Department ofBotany, University ofSheffield, Sheffield S1O 2TN, U.K. (Received 10 September 1975) Spinach chloroplast phosphoribulokinase is inhibited by DL-glyceraldehyde. The in- hibition is non-competitive with respect to ribulose 5-phosphate (Ki 19mM) and ATP (Ki 20mM). The inhibition is discussed in relation to a previously reported inhibition of CO2 assimilation in intact and envelope-free chloroplasts by DL-glyceraldehyde. It is concluded that the inhibition of phosphoribulokinase is insufficient to account for the inhibition, by DL-glyceraldehyde, of 02 evolution with ribose 5-phosphate as substrate and that a further site of inhibition is also present in this system. DL-Glyceraldehyde inhibits photosynthetic carbon glycylglycine buffer, pH7.4, in a total volume of assiniilation by intact chloroplasts and by the re- 13 ml. The reaction was terminated by the addition constituted chloroplast system (Stokes & Walker, of 2ml of 50 % (w/v) trichloroacetic acid and the pH 1972). The inhibition is most pronounced with intact was adjusted to pH8.0 with KOH. After the addition chloroplasts, a concentration of 1OmM being sufficient of Sml of 1.OM-barium acetate the solution was to suppress 02 evolution completely. It is important centrifuged and the precipitate discarded after because DL-glyceraldehyde is the only known inhibi- washing with Sml of water. Cold ethanol (4vol.) was tor of photosynthesis that is entirely without detect- added to the combined supernatants (total volume able effect on photophosphorylation or photo- 25.4ml); the new precipitate was recovered by synthetic electron transport. -

Structure-Function Studies of Enzymes from Ribose Metabolism
Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology 939 Structure-Function Studies of Enzymes from Ribose Metabolism BY C. EVALENA ANDERSSON ACTA UNIVERSITATIS UPSALIENSIS UPPSALA 2004 !"" #$"" % & % % ' ( ) * + &( , +( !""( - . - % + / % 0 ( , ( 1#1( ( ( 2-3 1. 45 ." 2 * & & * % * &( , % . * % % ( ) % / ( 0 6 / % ,)' & % % & ( )* % 6 % 6 * ( 0 6 * * % ( - % & 7 % & % & && ( ' && ,)' % /( 2 8 * ,)' & ,'.'' ( ) * % / % * 6 & & / 6 ( 0 . . . ( - * & * % %% & ( 9 * 6 / %% % ( -: % & * . & . , /( , & % * /( ) % / % & % ( ! 6 . . & / 6 % " # $ % # %& '()# %$# # *+',-. # ; ( + , !"" 2--3 ".!#!< 2-3 1. 45 ." $ $$$ .#111 = $>> (6(> ? @ $ $$$ .#111A List of Papers This thesis is based on the following papers, which are referred to in the text by their Roman numerals: I Andersson, C. E. & Mowbray, S. L. (2002). Activation of ribokinase by monovalent cations. J. Mol. Biol. 315, 409-19 II Zhang, R., Andersson, C. E., Savchenko, -

Swiveling Domain Mechanism in Pyruvate Phosphate Dikinase†,‡ Kap Lim,§ Randy J
Biochemistry 2007, 46, 14845-14853 14845 Swiveling Domain Mechanism in Pyruvate Phosphate Dikinase†,‡ Kap Lim,§ Randy J. Read,| Celia C. H. Chen,§ Aleksandra Tempczyk,§ Min Wei,⊥ Dongmei Ye,⊥ Chun Wu,⊥ Debra Dunaway-Mariano,⊥ and Osnat Herzberg*,§ Center for AdVanced Research in Biotechnology, UniVersity of Maryland Biotechnology Institute, RockVille, Maryland 20850, Department of Haematology, Cambridge Institute for Medical Research, UniVersity of Cambridge, Cambridge, United Kingdom, and Department of Chemistry, UniVersity of New Mexico, Albuquerque, New Mexico ReceiVed September 10, 2007; ReVised Manuscript ReceiVed October 17, 2007 ABSTRACT: Pyruvate phosphate dikinase (PPDK) catalyzes the reversible conversion of phosphoenolpyruvate (PEP), AMP, and Pi to pyruvate and ATP. The enzyme contains two remotely located reaction centers: the nucleotide partial reaction takes place at the N-terminal domain, and the PEP/pyruvate partial reaction takes place at the C-terminal domain. A central domain, tethered to the N- and C-terminal domains by two closely associated linkers, contains a phosphorylatable histidine residue (His455). The molecular architecture suggests a swiveling domain mechanism that shuttles a phosphoryl group between the two reaction centers. In an early structure of PPDK from Clostridium symbiosum, the His445-containing domain (His domain) was positioned close to the nucleotide binding domain and did not contact the PEP/pyruvate- binding domain. Here, we present the crystal structure of a second conformational state of C. symbiosum PPDK with the His domain adjacent to the PEP-binding domain. The structure was obtained by producing a three-residue mutant protein (R219E/E271R/S262D) that introduces repulsion between the His and nucleotide-binding domains but preserves viable interactions with the PEP/pyruvate-binding domain. -

Ceramides: Nutrient Signals That Drive Hepatosteatosis
J Lipid Atheroscler. 2020 Jan;9(1):50-65 Journal of https://doi.org/10.12997/jla.2020.9.1.50 Lipid and pISSN 2287-2892·eISSN 2288-2561 Atherosclerosis Review Ceramides: Nutrient Signals that Drive Hepatosteatosis Scott A. Summers Department of Nutrition and Integrative Physiology, University of Utah, Salt Lake City, UT, USA Received: Sep 24, 2019 ABSTRACT Revised: Nov 4, 2019 Accepted: Nov 10, 2019 Ceramides are minor components of the hepatic lipidome that have major effects on liver Correspondence to function. These products of lipid and protein metabolism accumulate when the energy needs Scott A. Summers of the hepatocyte have been met and its storage capacity is full, such that free fatty acids start Department of Nutrition and Integrative to couple to the sphingoid backbone rather than the glycerol moiety that is the scaffold for Physiology, University of Utah, 15N 2030E, Salt Lake City, UT 84112, USA. glycerolipids (e.g., triglycerides) or the carnitine moiety that shunts them into mitochondria. E-mail: [email protected] As ceramides accrue, they initiate actions that protect cells from acute increases in detergent- like fatty acids; for example, they alter cellular substrate preference from glucose to lipids Copyright © 2020 The Korean Society of Lipid and they enhance triglyceride storage. When prolonged, these ceramide actions cause insulin and Atherosclerosis. This is an Open Access article distributed resistance and hepatic steatosis, 2 of the underlying drivers of cardiometabolic diseases. under the terms of the Creative Commons Herein the author discusses the mechanisms linking ceramides to the development of insulin Attribution Non-Commercial License (https:// resistance, hepatosteatosis and resultant cardiometabolic disorders. -

Posttranslational Regulation of Pyruvate, Orthophosphate Dikinase in Developing Rice (Oryza Sativa) Seeds
Planta (2006) DOI 10.1007/s00425-006-0259-3 ORIGINAL ARTICLE Chris J. Chastain Æ Jarrod W. Heck Thomas A. Colquhoun Æ Dylan G. Voge Xing-You Gu Posttranslational regulation of pyruvate, orthophosphate dikinase in developing rice (Oryza sativa) seeds Received: 16 January 2006 / Accepted: 25 February 2006 Ó Springer-Verlag 2006 Abstract Pyruvate, orthophosphate dikinase (PPDK; (PPDK inactivation) and protein degradation. Immu- E.C.2.7.9.1) is most well known as a photosynthetic noblot analysis of separated seed tissue fractions enzyme in C4 plants. The enzyme is also ubiquitous in (pericarp, embryo + aleurone, seed embryo) revealed C3 plant tissues, although a precise non-photosynthetic that regulatory phosphorylation of PPDK occurs in the C3 function(s) is yet to be validated, owing largely to its non-green seed embryo and green outer pericarp layer, low abundance in most C3 organs. The single C3 organ but not in the endosperm + aleurone layer. The type where PPDK is in high abundance, and, therefore, modestly abundant pool of inactive PPDK (phosphor- where its function is most amenable to elucidation, are ylated + dephosphorylated) that was found to persist the developing seeds of graminaceous cereals. In this in mature rice seeds was shown to remain largely un- report, we suggest a non-photosynthetic function for C3 changed (inactive) upon seed germination, suggesting PPDK by characterizing its abundance and posttrans- that PPDK in rice seeds function in developmental lational regulation in developing Oryza sativa (rice) rather than in post-developmental processes. These and seeds. Using primarily an immunoblot-based approach, related observations lead us to postulate a putative we show that PPDK is a massively expressed protein function for the enzyme that aligns its PEP to pyruvate- during the early syncitial-endosperm/-cellularization forming reaction with biosynthetic processes that are stage of seed development.