Pyruvate Orthophosphate Dikinase
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hereditary Galactokinase Deficiency J
Arch Dis Child: first published as 10.1136/adc.46.248.465 on 1 August 1971. Downloaded from Alrchives of Disease in Childhood, 1971, 46, 465. Hereditary Galactokinase Deficiency J. G. H. COOK, N. A. DON, and TREVOR P. MANN From the Royal Alexandra Hospital for Sick Children, Brighton, Sussex Cook, J. G. H., Don, N. A., and Mann, T. P. (1971). Archives of Disease in Childhood, 46, 465. Hereditary galactokinase deficiency. A baby with galactokinase deficiency, a recessive inborn error of galactose metabolism, is des- cribed. The case is exceptional in that there was no evidence of gypsy blood in the family concerned. The investigation of neonatal hyperbilirubinaemia led to the discovery of galactosuria. As noted by others, the paucity of presenting features makes early diagnosis difficult, and detection by biochemical screening seems desirable. Cataract formation, of early onset, appears to be the only severe persisting complication and may be due to the biosynthesis and accumulation of galactitol in the lens. Ophthalmic surgeons need to be aware of this enzyme defect, because with early diagnosis and dietary treatment these lens changes should be reversible. Galactokinase catalyses the conversion of galac- and galactose diabetes had been made in this tose to galactose-l-phosphate, the first of three patient (Fanconi, 1933). In adulthood he was steps in the pathway by which galactose is converted found to have glycosuria as well as galactosuria, and copyright. to glucose (Fig.). an unexpectedly high level of urinary galactitol was detected. He was of average intelligence, and his handicaps, apart from poor vision, appeared to be (1) Galactose Gackinase Galactose-I-phosphate due to neurofibromatosis. -

Biochemical and Comparative Transcriptome Analyses Reveal
biomolecules Article Biochemical and Comparative Transcriptome Analyses Reveal Key Genes Involved in Major Metabolic Regulation Related to Colored Leaf Formation in Osmanthus fragrans ‘Yinbi Shuanghui’ during Development Xuan Chen 1,2, Xiulian Yang 1,3, Jun Xie 2, Wenjie Ding 1,3, Yuli Li 1,3, Yuanzheng Yue 1,3,* and Lianggui Wang 1,3,* 1 Key Laboratory of Landscape Architecture, Jiangsu Province, College of Landscape Architecture, Nanjing Forestry University, No. 159 Longpan Road, Nanjing 210037, China; [email protected] (X.C.); [email protected] (X.Y.); [email protected] (W.D.); [email protected] (Y.L.) 2 College of Fine Arts, Nanjing Normal University of Special Education, No.1 Shennong Road, Nanjing 210038, China; [email protected] 3 Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing 210037, China * Correspondence: [email protected] (Y.Y.); [email protected] (L.W.); Tel.: +86-138-0900-7625 (L.W.) Received: 25 February 2020; Accepted: 1 April 2020; Published: 4 April 2020 Abstract: Osmanthus fragrans ‘Yinbi Shuanghui’ not only has a beautiful shape and fresh floral fragrance, but also rich leaf colors that change, making the tree useful for landscaping. In order to study the mechanisms of color formation in O. fragrans ‘Yinbi Shuanghui’ leaves, we analyzed the colored and green leaves at different developmental stages in terms of leaf pigment content, cell structure, and transcriptome data. We found that the chlorophyll content in the colored leaves was lower than that of green leaves throughout development. By analyzing the structure of chloroplasts, the colored leaves demonstrated more stromal lamellae and low numbers of granum thylakoid. -

Induction of Uridyl Transferase Mrna-And Dependency on GAL4 Gene Function (In Vitro Translation/Immunoprecipitation/GAL Gene Cluster/Positive Regulation) JAMES E
Proc. Nati. Acad. Sci. USA Vol. 75, No. 6, pp. 2878-2882, June 1978 Genetics Regulation of the galactose pathway in Saccharomyces cerevisiae: Induction of uridyl transferase mRNA-and dependency on GAL4 gene function (in vitro translation/immunoprecipitation/GAL gene cluster/positive regulation) JAMES E. HOPPER*, JAMES R. BROACHt, AND LUCY B. ROWE* * Rosenstiel Basic Medical Sciences Research Center, Brandeis University, Waltham, Massachusetts 02154; and t Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 11724 Communicated by Norman H. Giles, April 10,1978 ABSTRACT In Saccharomyces cerevisiae, utilization of Genetic control of the inducible galactose pathway enzymes galactose requires four inducible enzyme activities. Three of involves the four structural genes GALI, GAL10, GAL7, and these activities (galactose-l-phosphate uridyl transferase, EC genes, GAL4, GAL81 (c), GAL80 2.7.7.10; uridine diphosphogalactose 4-epimerase, EC 5.1.3.2; GAL2 and four regulatory and galactokinase, EC 2.7.1.6) are specified by three tightly (i), and GALS.* Mutations in GALl, GAL10, GAL7, and GAL2 linked genes (GAL7, GALlO, and GALI, respectively) on chro- affect the individual appearance of galactokinase, epimerase, mosome II, whereas the fourth, galactose transport, is specified transferase, and galactose transport activities, respectively (6). by a gene (GALS) located on chromosome XIL Although classic Mutations defining the GALl, GAL10, and GAL7 genes have genetic analysis has revealed both positive and negative regu- invariably been recessive, and they map in three tightly linked latory genes that coordinately affect the appearance of ail four complementation groups near the centromere of chromosome enzyme activities, neither the basic events leading to the ap- pearance of enzyme activities nor the roles of the regulatory II (6, 9, 10). -

Open Full Page
Published OnlineFirst February 12, 2018; DOI: 10.1158/0008-5472.CAN-17-2215 Cancer Metabolism and Chemical Biology Research RSK Regulates PFK-2 Activity to Promote Metabolic Rewiring in Melanoma Thibault Houles1, Simon-Pierre Gravel2,Genevieve Lavoie1, Sejeong Shin3, Mathilde Savall1, Antoine Meant 1, Benoit Grondin1, Louis Gaboury1,4, Sang-Oh Yoon3, Julie St-Pierre2, and Philippe P. Roux1,4 Abstract Metabolic reprogramming is a hallmark of cancer that includes glycolytic flux in melanoma cells, suggesting an important role for increased glucose uptake and accelerated aerobic glycolysis. This RSK in BRAF-mediated metabolic rewiring. Consistent with this, phenotypeisrequiredtofulfill anabolic demands associated with expression of a phosphorylation-deficient mutant of PFKFB2 aberrant cell proliferation and is often mediated by oncogenic decreased aerobic glycolysis and reduced the growth of melanoma drivers such as activated BRAF. In this study, we show that the in mice. Together, these results indicate that RSK-mediated phos- MAPK-activated p90 ribosomal S6 kinase (RSK) is necessary to phorylation of PFKFB2 plays a key role in the metabolism and maintain glycolytic metabolism in BRAF-mutated melanoma growth of BRAF-mutated melanomas. cells. RSK directly phosphorylated the regulatory domain of Significance: RSK promotes glycolytic metabolism and the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 (PFKFB2), growth of BRAF-mutated melanoma by driving phosphory- an enzyme that catalyzes the synthesis of fructose-2,6-bisphosphate lation of an important glycolytic enzyme. Cancer Res; 78(9); during glycolysis. Inhibition of RSK reduced PFKFB2 activity and 2191–204. Ó2018 AACR. Introduction but recently developed therapies that target components of the MAPK pathway have demonstrated survival advantage in pati- Melanoma is the most aggressive form of skin cancer and arises ents with BRAF-mutated tumors (7). -
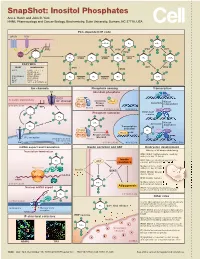
Snapshot: Inositol Phosphates Ace J
SnapShot: Inositol Phosphates Ace J. Hatch and John D. York HHMI, Pharmacology and Cancer Biology, Biochemistry, Duke University, Durham, NC 27710, USA PLC-dependent IP code GPCR RTK O O O O O O 5-PP-IP4 IP4 5-IP7 O O O O O O PIP2 O IP6K O IP6K O VIP1 O O O 2 O ITPK1 O 13 O PLC 2 O O O O O O O O 4 6 13 O 5 IP3 IPMK IP4 IPMK IP5 IPK1 IP6 1,5-IP8 4 6 O O 5 O O O O O O O O ENZYMES O O O O O O YEAST MAMMALIAN IP3K VIP1 IP6K IPMK PLC1 PLCβ, γ, δ, ε, ζ, η O - IP3KA, B, C - ITPK1 (IP56K) O O O O O O O O IPK2(ARG82) IPMK (IPK2) IP4 IP3 IP4 1-IP7 IPK1 IPK1 (IP5K) INPP5 ITPK1 KCS1 IP6K1, 2, 3 O O O O O O VIP1 VIP1, 2 (PPIP5K1, 2) O O Ion channels Phosphate sensing Transcription Cl- Abundant phosphate MCM1 ARG80 CIC3 P PLASMA MEMBRANE - Pho80 Cl channel Pho4 Kinase Kinase Assembly Pho85 independent CYTOPLASM activity 2 O PIP2 Pho81 13 CYTOPLASM NUCLEUS IPK2 ARG81 4 6 Phosphate starvation MCM1-ArgR O 5 O complex O O IP4 O O O O O O O 1-IP7 Kinase Activation dependent IP3 O O Transcription O O O activated Pho80 IP4 O X Pho4 O O Pho85 Kinase activity IP receptor blocked O 3 ENDOPLASMIC Pho81 RETICULUM Ca2+ CYTOPLASM NUCLEUS NUCLEUS mRNA export and translation Insulin secretion and AKT Embryonic development Translation termination Effects of IP kinase deficiency O IPMK (IPK2): Multiple defects, death by embryonic day 10 (mice) O O Insulin IPK1: Cillia are shortened and immotile IP6 AKT resistance causing patterning defects (zebrash) O O Multiple defects, death by Ribosome O embryonic day 8.5 (mice) GleI eRF1 Insulin GSK3β Dbp5 ITPK1 (IP56K): Neural tube -

Effect of Enzyme/Substrate Ratio on the Antioxidant Properties of Hydrolysed African Yam Bean
African Journal of Biotechnology Vol. 11(50), pp. 11086-11091, 21 June, 2012 Available online at http://www.academicjournals.org/AJB DOI: 10.5897/AJB11.2271 ISSN 1684–5315 ©2012 Academic Journals Full Length Research Paper Effect of enzyme/substrate ratio on the antioxidant properties of hydrolysed African yam bean Fasasi Olufunmilayo*, Oyebode Esther and Fagbamila Oluwatoyin Department of Food Science and Technology, P. M. B. 704, Federal University of Technology, Akure, Nigeria. Accepted 8 June, 2012 The use of natural antioxidant as compared with synthetic antioxidant in food processing is a growing trend as consumers prefer natural to synthetic antioxidant mainly on emotional ground. This study investigates the antioxidant activity of hydrolysed African yam bean (Sphenostylis sternocarpa) which is regarded as one of the neglected underutilized species (NUS) of crop in Africa and Nigeria especially to improve food security and boost the economic importance of the crop. The antioxidant properties of African yam bean hydrolysates (AYH) produced at different enzyme to substrate (E/S) ratios of 1: 100 and 3: 100 (W/V) using pepsin (pH 2.0, 37°C) were studied. 2, 2-Diphenyl-1-picryl-hydrazyl (DPPH) radical scavenging activity of the hydrolysates was significantly influenced by the E\S ratio as DPPH radical scavenging activity ranged from 56.1 to 75.8% in AYH (1: 100) and 33.3 to 58.8% in AYH (3:100) with 1000 µg/ml having the highest DPPH radical scavenging activity. AYH (1:100) and AYH (3:100) had higher reducing activities of 0.42 and 0.23, respectively at a concentration of 1000 µg/ml. -

Role of Glucokinase and Glucose-6 Phosphatase in the Nutritional Regulation of Endogenous Glucose Production G Mithieux
Role of glucokinase and glucose-6 phosphatase in the nutritional regulation of endogenous glucose production G Mithieux To cite this version: G Mithieux. Role of glucokinase and glucose-6 phosphatase in the nutritional regulation of endogenous glucose production. Reproduction Nutrition Development, EDP Sciences, 1996, 36 (4), pp.357-362. hal-00899845 HAL Id: hal-00899845 https://hal.archives-ouvertes.fr/hal-00899845 Submitted on 1 Jan 1996 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Review Role of glucokinase and glucose-6 phosphatase in the nutritional regulation of endogenous glucose production G Mithieux Unité 197 de l’Inserm, faculté de médecine René-Laënnec, rue Guillaume-Paradin, 69372 Lyon cedex 08, France (Received 29 November 1995; accepted 6 May 1996) Summary ― Two specific enzymes, glucokinase (GK) and glucose-6 phosphatase (Gic6Pase) enable the liver to play a crucial role in glucose homeostasis. The activity of Glc6Pase, which enables the liver to produce glucose, is increased during short-term fasting, in agreement with the enhancement of liver gluconeogenesis. During long-term fasting, Glc6Pase activity is increased in the kidney, which con- tributes significantly to the glucose supply at that time. -

Regulation of Energy Substrate Metabolism in Endurance Exercise
International Journal of Environmental Research and Public Health Review Regulation of Energy Substrate Metabolism in Endurance Exercise Abdullah F. Alghannam 1,* , Mazen M. Ghaith 2 and Maha H. Alhussain 3 1 Lifestyle and Health Research Center, Health Sciences Research Center, Princess Nourah bInt. Abdulrahman University, Riyadh 84428, Saudi Arabia 2 Faculty of Applied Medical Sciences, Laboratory Medicine Department, Umm Al-Qura University, Al Abdeyah, Makkah 7607, Saudi Arabia; [email protected] 3 Department of Food Science and Nutrition, College of Food and Agriculture Sciences, King Saud University, Riyadh 11451, Saudi Arabia; [email protected] * Correspondence: [email protected] Abstract: The human body requires energy to function. Adenosine triphosphate (ATP) is the cellular currency for energy-requiring processes including mechanical work (i.e., exercise). ATP used by the cells is ultimately derived from the catabolism of energy substrate molecules—carbohydrates, fat, and protein. In prolonged moderate to high-intensity exercise, there is a delicate interplay between carbohydrate and fat metabolism, and this bioenergetic process is tightly regulated by numerous physiological, nutritional, and environmental factors such as exercise intensity and du- ration, body mass and feeding state. Carbohydrate metabolism is of critical importance during prolonged endurance-type exercise, reflecting the physiological need to regulate glucose homeostasis, assuring optimal glycogen storage, proper muscle fuelling, and delaying the onset of fatigue. Fat metabolism represents a sustainable source of energy to meet energy demands and preserve the ‘limited’ carbohydrate stores. Coordinated neural, hormonal and circulatory events occur during prolonged endurance-type exercise, facilitating the delivery of fatty acids from adipose tissue to the Citation: Alghannam, A.F.; Ghaith, working muscle for oxidation. -

Terminal Deoxynucleotidyl Transferase Protocol
Certificate of Analysis Terminal Deoxynucleotidyl Transferase, Recombinant: Part No. Size (units) Part# 9PIM187 M828A 300 Revised 4/18 M828C 1,500 Description: This enzyme catalyzes the repetitive addition of mononucleotides to the terminal 3´-OH of a DNA initiator accompanied by the release of inorganic phosphate. Single-stranded DNA is preferred as an initiator. Polymerization is not template-dependent. The addition of 1mM Co2+ (as CoCl2) in the reaction buffer allows the tailing of 3´-ends with varying degrees of efficiency. Enzyme Storage Buffer: Terminal Deoxynucleotidyl Transferase, Recombinant, is supplied in 50mM potassium phosphate *AF9PIM187 0418M187* (pH 6.4), 100mM NaCl, 1mM β-mercaptoethanol, 0.1% Tween® 20 and 50% glycerol. AF9PIM187 0418M187 Source: Recombinant E. coli strain. Storage Conditions: See the Product Information Label for storage recommendations. Avoid multiple freeze-thaw cycles and exposure to frequent temperature changes. See the expiration date on the Product Information Label. Terminal Transferase 5X Buffer (M189A): 500mM cacodylate buffer (pH 6.8), 5mM CoCl2 and 0.5mM DTT. Unit Definition: One unit of activity catalyzes the transfer of 0.5 picomoles of ddATP to oligo(dT)16 per minute at 37°C in 1X Terminal Transferase Buffer. The resulting oligo(dT)17 is measured by HPLC. Usage Notes for 3´-End Labeling Reaction 1. Not all dNTPs are tailed with the same efficiency. Actual concentration of dNTP will depend on the individual application. 2. The provided buffer (5X) is to be used in the tailing reaction. The recommended reaction conditions are as described under Quality Control Assays, 3´-End Labeling Reaction, and in Section III overleaf. -

Commentary Tracking Telomerase
Cell, Vol. S116, S83–S86, January 23, 2004 Copyright 2004 by Cell Press Tracking Telomerase Commentary Carol W. Greider1 and Elizabeth H. Blackburn2,* neous size of the fragments in gel electrophoresis was 1Department of Molecular Biology and Genetics the first suggestion of unusual behavior of telomeric Johns Hopkins University School of Medicine DNA. A similar telomere repeat sequence, CCCCAAAA 725 North Wolfe Street was soon found on natural chromosome ends in other Baltimore, Maryland 21205 ciliates (Klobutcher et al., 1981). Another very unusual 2 Department of Biochemistry and Biophysics finding regarding these repeated sequences came in University of California, San Francisco 1982: David Prescott found that these repeated sequences Box 2200 are added de novo to ciliate chromosomes during the San Francisco, California 94143 developmental process of chromosome fragmentation (Boswell et al., 1982). This was the first hint that a special mechanism may exist to add telomere repeats. The Telomere Problem The next clue came from work in yeast. In a remarkable The paper reprinted here, the initial identification of telo- example of functional conservation across phylogenetic merase, resulted from our testing a very specific hypoth- kingdoms, Liz and Jack Szostak (Szostak and Black- esis: that an enzyme existed, then undiscovered, that burn, 1982) showed that the Tetrahymena telomeric se- could add telomeric repeats onto chromosome ends. quences could replace the yeast telomere entirely. A We based this hypothesis on several unexplained facts mini-chromosome with these foreign telomeres main- and creative questions being asked by people who were tained its linear structure and replicated and segregated trying to understand those facts. -

Ceramide Kinase Is Upregulated in Metastatic Breast Cancer Cells and Contributes to Migration and Invasion by Activation of PI 3-Kinase and Akt
International Journal of Molecular Sciences Article Ceramide Kinase Is Upregulated in Metastatic Breast Cancer Cells and Contributes to Migration and Invasion by Activation of PI 3-Kinase and Akt 1,2, 1, 2 2 Stephanie Schwalm y, Martin Erhardt y, Isolde Römer , Josef Pfeilschifter , Uwe Zangemeister-Wittke 1,* and Andrea Huwiler 1,* 1 Institute of Pharmacology, University of Bern, Inselspital, INO-F, CH-3010 Bern, Switzerland; [email protected] (S.S.); [email protected] (M.E.) 2 Institute of General Pharmacology and Toxicology, University Hospital Frankfurt am Main, Goethe-University, Theodor-Stern Kai 7, D-60590 Frankfurt am Main, Germany; [email protected] (I.R.); [email protected] (J.P.) * Correspondence: [email protected] (U.Z.-W.); [email protected] (A.H.); Tel.: +41-31-6323214 (A.H.) These authors contributed equally to this work. y Received: 29 November 2019; Accepted: 12 February 2020; Published: 19 February 2020 Abstract: Ceramide kinase (CerK) is a lipid kinase that converts the proapoptotic ceramide to ceramide 1-phosphate, which has been proposed to have pro-malignant properties and regulate cell responses such as proliferation, migration, and inflammation. We used the parental human breast cancer cell line MDA-MB-231 and two single cell progenies derived from lung and bone metastasis upon injection of the parental cells into immuno-deficient mice. The lung and the bone metastatic cell lines showed a marked upregulation of CerK mRNA and activity when compared to the parental cell line. The metastatic cells also had increased migratory and invasive activity, which was dose-dependently reduced by the selective CerK inhibitor NVP-231. -

Allosteric Regulation
Hanjia’s Biochemistry Lecture Hanjia’s Biochemistry Lecture Chapter 15 Essential Questions • Before this class, ask your self the following questions: Reginald H. Garrett – What are the properties of regulatory enzymes? Enzyygme Regulation Charles M. Grisham • How do you know this enzyme is a regulatory enzyme? – How do regulatory enzymes sense the momentary needfll?ds of cells? ?Դৣ • How signal is delivered ᑣ٫݅ ፐำᆛઠ – Wha t mo lecu lar mec han isms are used to regu la te enzyme activity? http://lms. ls. ntou. edu. tw/course/106 [email protected] 2 Hanjia’s Biochemistry Lecture Hanjia’s Biochemistry Lecture Outline 15. 1 – What Factors Influence Enzymatic Activity? • Part 1 Factors that influence enzymatic activity 1. The availability of substrates and cofactors! – Zymogen, isozyme and covalent modification! 2. Product accu m ul ates b the rate will dec r ease! • Part 2: The general features of allosteric 3. The amount of enzyme present at any moment – Genetic regulation of enzyme synthesis and decay regulation 4. Regulation of Enzyme activity – The mechanisms of allosteric regulation – Zymogens, isozymes , and modulator proteins may play – Example of a enzyme controlled by both a role allosteric regulation and covalent modification – Enzyme activity can be regulated through covalent modification • Part 3: Special focus on hemoglobin and – Allosteric Regulation myoglbilobin 3 4 Hanjia’s Biochemistry Lecture Hanjia’s Biochemistry Lecture Regulation 1: Zymogen … The proteolytic activation of chymotrypsinogen • Zymogens are inactive