Staphylococcus Aureus Targets the Purine Salvage Pathway to Kill Phagocytes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Supplementary Materials
1 Supplementary Materials: Supplemental Figure 1. Gene expression profiles of kidneys in the Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice. (A) A heat map of microarray data show the genes that significantly changed up to 2 fold compared between Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice (N=4 mice per group; p<0.05). Data show in log2 (sample/wild-type). 2 Supplemental Figure 2. Sting signaling is essential for immuno-phenotypes of the Fcgr2b-/-lupus mice. (A-C) Flow cytometry analysis of splenocytes isolated from wild-type, Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice at the age of 6-7 months (N= 13-14 per group). Data shown in the percentage of (A) CD4+ ICOS+ cells, (B) B220+ I-Ab+ cells and (C) CD138+ cells. Data show as mean ± SEM (*p < 0.05, **p<0.01 and ***p<0.001). 3 Supplemental Figure 3. Phenotypes of Sting activated dendritic cells. (A) Representative of western blot analysis from immunoprecipitation with Sting of Fcgr2b-/- mice (N= 4). The band was shown in STING protein of activated BMDC with DMXAA at 0, 3 and 6 hr. and phosphorylation of STING at Ser357. (B) Mass spectra of phosphorylation of STING at Ser357 of activated BMDC from Fcgr2b-/- mice after stimulated with DMXAA for 3 hour and followed by immunoprecipitation with STING. (C) Sting-activated BMDC were co-cultured with LYN inhibitor PP2 and analyzed by flow cytometry, which showed the mean fluorescence intensity (MFI) of IAb expressing DC (N = 3 mice per group). 4 Supplemental Table 1. Lists of up and down of regulated proteins Accession No. -

Novocib Nucleoside Kinases
PRECICE ® Services Information sheet Nucleoside kinases, rate-limiting step of nucleoside analogues activation Nucleoside analogues have proven to be a highly successful class of anti-cancer and anti-viral drugs. The therapeutic efficacy of nucleoside analogues is dependent of their intracellular phosphorylation. Two cellular nucleoside kinases, deoxycytidine kinase (dCK) and UMP-CMP kinase (CMK) are critical for phosphorylation of cytidine analogues. These kinases provide two first steps of activation of highly effective anti-cancer and anti-viral drugs, such as 1-β-D- arabinofuranosylcytosine (araC, aracytidine ), 2’,2’difluorodeoxycytidine (dFdC, gemcitabine ), β-D-2’3’- dideoxycytidine (ddC). Both kinases phosphorylate unnatural L-nucleosides (e.g., β-L-2’3’-dideoxy-3’thiacytidine, L- SSdC, 3-TC or lamividune ). Kinetic constants of araC, dFdC and 3TC phosphorylation by recombinant dCK and UMP-CMPK have been published. The comparison of phosphorylation properties of new nucleoside analogues with those of known drugs provides the rational basis for selection of analogues of better therapeutic potential. To characterize the phosphorylation properties of new nucleoside analogues, Novo CIB has developed human recombinant dCK and human recombinant CMK nucleoside phosphorylation assays. As shown in Table 1, CMK assay must be performed with monophosphate forms of nucleoside analogues and requires preliminary phosphorylation of nucleoside analogues and their purification. To circumvent this time-consuming step, NovoCIB has developed a coupled dCK-CMK nucleoside phosphorylation assay that delivers in one step the critical information on both dCK and CMK substrate properties of nucleoside analogue. Ribavirin (1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide) is a purine nucleoside analogue with a broad-spectrum antiviral activity. -

Ribonucleotide Reductase Messenger RNA Expression and Survival in Gemcitabine/Cisplatin-Treated Advanced Non-Small Cell Lung Cancer Patients
1318 Vol. 10, 1318–1325, February 15, 2004 Clinical Cancer Research Ribonucleotide Reductase Messenger RNA Expression and Survival in Gemcitabine/Cisplatin-Treated Advanced Non-Small Cell Lung Cancer Patients Rafael Rosell,1 Kathleen D. Danenberg,2 from bronchoscopy by real-time quantitative reverse tran- Vincente Alberola,3 Gerold Bepler,4 scription-PCR. Results were correlated with survival using Jose Javier Sanchez,5 Carlos Camps,6 the Kaplan-Meier method. 7 8 Results: A total of 100 patients were assessed. There was Mariano Provencio, Dolores Isla, a strong correlation between RRM1 and ERCC1 mRNA 1 9 P < 0.001). In the ;0.410 ؍ Miquel Taron, Pilar Diz, and expression levels (Spearman r Angel Artal10 on behalf of the Spanish Lung gemcitabine/cisplatin arm, patients with low RRM1 mRNA Cancer Group expression levels had significantly longer median survival 1Medical Oncology Service, Institut Catala`d’Oncologia, Hospital than those with high levels [13.7 versus 3.6 months; 95% -Me .[0.009 ؍ Germans Trias i Pujol, Barcelona, Spain; 2Response Genetics Inc, confidence interval (CI), 9.6–17.8 months; P Los Angeles, California; 3Medical Oncology Service, Hospital Arnau dian survival was also significantly longer among patients 4 de Vilanova, Valencia, Spain; Thoracic Oncology Program, H. Lee with low mRNA expression levels of both RRM1 and ERCC1 Moffitt Cancer Center and Research Institute, Tampa, Florida; 5 (not reached), than among those with high levels of both ؍ ,Facultat de Medicina, Autonomous University of Madrid, Madrid Spain; 6Medical Oncology Service, Hospital General de Valencia, genes (6.8 months; 95% CI, 2.6–11.1 months; P 0.016). -

Supplementary File 2A Revised
Supplementary file 2A. Differentially expressed genes in aldosteronomas compared to all other samples, ranked according to statistical significance. Missing values were not allowed in aldosteronomas, but to a maximum of five in the other samples. Acc UGCluster Name Symbol log Fold Change P - Value Adj. P-Value B R99527 Hs.8162 Hypothetical protein MGC39372 MGC39372 2,17 6,3E-09 5,1E-05 10,2 AA398335 Hs.10414 Kelch domain containing 8A KLHDC8A 2,26 1,2E-08 5,1E-05 9,56 AA441933 Hs.519075 Leiomodin 1 (smooth muscle) LMOD1 2,33 1,3E-08 5,1E-05 9,54 AA630120 Hs.78781 Vascular endothelial growth factor B VEGFB 1,24 1,1E-07 2,9E-04 7,59 R07846 Data not found 3,71 1,2E-07 2,9E-04 7,49 W92795 Hs.434386 Hypothetical protein LOC201229 LOC201229 1,55 2,0E-07 4,0E-04 7,03 AA454564 Hs.323396 Family with sequence similarity 54, member B FAM54B 1,25 3,0E-07 5,2E-04 6,65 AA775249 Hs.513633 G protein-coupled receptor 56 GPR56 -1,63 4,3E-07 6,4E-04 6,33 AA012822 Hs.713814 Oxysterol bining protein OSBP 1,35 5,3E-07 7,1E-04 6,14 R45592 Hs.655271 Regulating synaptic membrane exocytosis 2 RIMS2 2,51 5,9E-07 7,1E-04 6,04 AA282936 Hs.240 M-phase phosphoprotein 1 MPHOSPH -1,40 8,1E-07 8,9E-04 5,74 N34945 Hs.234898 Acetyl-Coenzyme A carboxylase beta ACACB 0,87 9,7E-07 9,8E-04 5,58 R07322 Hs.464137 Acyl-Coenzyme A oxidase 1, palmitoyl ACOX1 0,82 1,3E-06 1,2E-03 5,35 R77144 Hs.488835 Transmembrane protein 120A TMEM120A 1,55 1,7E-06 1,4E-03 5,07 H68542 Hs.420009 Transcribed locus 1,07 1,7E-06 1,4E-03 5,06 AA410184 Hs.696454 PBX/knotted 1 homeobox 2 PKNOX2 1,78 2,0E-06 -

The KMT1A-GATA3-STAT3 Circuit Is a Novel Self-Renewal Signaling of Human Bladder Cancer Stem Cells Zhao Yang1, Luyun He2,3, Kais
The KMT1A-GATA3-STAT3 circuit is a novel self-renewal signaling of human bladder cancer stem cells Zhao Yang1, Luyun He2,3, Kaisu Lin4, Yun Zhang1, Aihua Deng1, Yong Liang1, Chong Li2, 5, & Tingyi Wen1, 6, 1CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China 2Core Facility for Protein Research, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China 3CAS Key Laboratory of Infection and Immunity, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China 4Department of Oncology, the Second Affiliated Hospital of Soochow University, Suzhou 215000, China 5Beijing Jianlan Institute of Medicine, Beijing 100190, China 6Savaid Medical School, University of Chinese Academy of Sciences, Beijing 100049, China Correspondence author: Tingyi Wen, e-mail: [email protected] Chong Li, e-mail: [email protected] Supplementary Figure S1. Isolation of human bladder cancer stem cells. BCMab1 and CD44 were used to isolate bladder cancer stem cells (BCSCs: BCMab1+CD44+) and bladder cancer non-stem cells (BCNSCs: BCMab1-CD44-) from EJ, samples #1 and #2 by flow cytometry. Supplementary Figure S2. Gene ontology analysis of downregulated genes of human BCSCs. (A) Pathway enrichment of 103 downregulated genes in BCSCs. (B) The seven downregulated genes in BCSCs participating in centromeric heterochromatin, mRNA-3’-UTR binding and translation regulator activity signaling pathways were validated by qRT-PCR. Data are presented as mean ± SD. P < 0.05; P < 0.01. Supplementary Figure S3. The expression of KMT1A is higher in human BC than that in peri-tumor tissues. (A) The expression of KMT1A was higher in BC samples than that in peri-tumors as assessed by immunohistochemistry, Scale bar = 50 m. -

The Effect of the Polyadenylation Inhibitor Cordycepin on MCF-7 Cells
The effect of the polyadenylation inhibitor Cordycepin on MCF-7 cells Asma Khurshid, MSc (University of Nottingham) Thesis submitted to the University of Nottingham for the degree of Doctor of Philosophy July 2015 Declaration Except where acknowledged in the text, I declare that this dissertation is my own work and is based on research that was undertaken by me in the School of Pharmacy, Faculty of Science, University of Nottingham. i Abstract Cordycepin (3′-deoxyadenosine) is a medicinal bioactive component of the caterpillar fungi (Cordyceps and Ophicordyceps). It is reported to have nephroprotective, antiapoptotic, anti-metastatic, hepatoprotective (Yue et al. 2013), inflammatory effects, antioxidant, anti-tumor, immunomodulatory and vasorelaxation activities. Cordycepin is well known to terminate and inhibit polyadenylation, both in vitro and in vivo. Other proposed mechanisms of action of cordycepin include activation of adenosine receptors, activation of AMP dependent kinase (AMPK) and inhibition of PARP1. The purpose of this study is to elucidate the biological and pharmacological effects of cordycepin on cancer cell lines such as MCF-7 cells. In this study I found that cordycepin reduces the cell proliferation in all examined cell lines without always exerting an effect on 4EBP phosphorylation and protein synthesis rates. Therefore, the effects on protein synthesis via inhibition of mTOR, which were previously reported, are not only the sole reason for the effect of cordycepin on cell proliferation. Knockdown of poly (A) polymerases reduces cell proliferation and survival, indicating that poly (A) polymerases are potential targets of cordycepin. I studied different adenosine analogues and found that 8 aminoadenosine, the only one that also consistently inhibits polyadenylation, also reduces levels of P-4EBP. -

The Genetic Program of Pancreatic Beta-Cell Replication in Vivo
Page 1 of 65 Diabetes The genetic program of pancreatic beta-cell replication in vivo Agnes Klochendler1, Inbal Caspi2, Noa Corem1, Maya Moran3, Oriel Friedlich1, Sharona Elgavish4, Yuval Nevo4, Aharon Helman1, Benjamin Glaser5, Amir Eden3, Shalev Itzkovitz2, Yuval Dor1,* 1Department of Developmental Biology and Cancer Research, The Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 2Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel. 3Department of Cell and Developmental Biology, The Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem 91904, Israel 4Info-CORE, Bioinformatics Unit of the I-CORE Computation Center, The Hebrew University and Hadassah, The Institute for Medical Research Israel- Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 5Endocrinology and Metabolism Service, Department of Internal Medicine, Hadassah-Hebrew University Medical Center, Jerusalem 91120, Israel *Correspondence: [email protected] Running title: The genetic program of pancreatic β-cell replication 1 Diabetes Publish Ahead of Print, published online March 18, 2016 Diabetes Page 2 of 65 Abstract The molecular program underlying infrequent replication of pancreatic beta- cells remains largely inaccessible. Using transgenic mice expressing GFP in cycling cells we sorted live, replicating beta-cells and determined their transcriptome. Replicating beta-cells upregulate hundreds of proliferation- related genes, along with many novel putative cell cycle components. Strikingly, genes involved in beta-cell functions, namely glucose sensing and insulin secretion were repressed. Further studies using single molecule RNA in situ hybridization revealed that in fact, replicating beta-cells double the amount of RNA for most genes, but this upregulation excludes genes involved in beta-cell function. -

Cytosine Arabinoside Transport and Metabolism in Acute Leukemias and T Cell Lymphoblastic Lymphoma
Cytosine arabinoside transport and metabolism in acute leukemias and T cell lymphoblastic lymphoma. J S Wiley, … , W H Sawyer, L R Finch J Clin Invest. 1985;75(2):632-642. https://doi.org/10.1172/JCI111741. Research Article Cytosine arabinoside (araC) has proven efficacy in acute myeloid leukemia (AML), but its place in the treatment of acute lymphoblastic leukemia (ALL) and T lymphoblastic lymphoma is uncertain. The therapeutic potential of araC has been assessed in patients with AML, ALL, and T lymphoblastic lymphoma by measuring the conversion of araC to its active metabolite, the 5'-triphosphate of araC (araCTP), in purified blasts from patients as well as in normal polymorphs and lymphocytes. In all leukemias, araCTP was the major intracellular metabolite of araC. The highest araCTP formation was in blasts from T lymphoblastic lymphoma, which formed threefold more nucleotide than myeloblasts, and in turn myeloblasts formed twofold more araCTP than lymphoblasts from ALL. The mean araCTP formation in myeloblasts was sixfold greater than polymorphs, but in contrast, lymphoblasts and lymphocytes formed low and similar amounts of this nucleotide. Reasons for the sixfold range in araCTP accumulation in the various leukemic blasts were studied. The mean size of myeloblasts was 35-70% larger than lymphoblasts when compared on the basis of protein or intracellular water content, but T lymphoblastic lymphoma blasts and lymphoblasts were the same size. Activities of deoxycytidine kinase, deoxycytidylate deaminase, and pyrimidine nucleoside monophosphate kinase were not different between any of the leukemic cell types. The number of nucleoside transport sites on blasts was estimated by measuring the […] Find the latest version: https://jci.me/111741/pdf Cytosine Arabinoside Transport and Metabolism in Acute Leukemias and T Cell Lymphoblastic Lymphoma J. -

Supplementary Table 1
Up-regulated accession # Development M93275 ADFP, adipose differentiation related protein D43694 MATH-1, homolog of atonal 1 M64068 Bmi-1, zinc finger protein AW124785 Midnolin, midbrain nucleolar protein AI843178 Cla3, Cerebellar ataxia 3 D10712 Nedd1, Neural precursor cell expressed, developmentally down-regulated gene 1 AB011678 Doublecortin, for neurogenesis M57683 mPDGF-alpha-R, PDGF alpha receptor U41626 DSS1, deleted in split hand/split foot 1 homolog (Dss1), for limb development AB010833 PTCH2, patched 2, Mouse homolog of yeast CDC46 NP_034226 Ebf3, early B-cell factor 3 AI846695 Qk, Quaking U63386 Edr1 Early development regulator 1 (homolog of polyhomeotic 1), Mph1 AI043016 Rnf2, Ring finger protein 2 X69942 Enhancer-trap-locus 1, for transcription regulation AF100694 Ruvbl1, Ruv-B like protein 1, DNA helicase AW123618 Fzd2, Frizzled homolog 2 U88566 Sfrp1, secreted frizzled related protein 1 AA681520 Geminin-pending, for embryogenesis and morphogenesis U88567 Sfrp2, secreted frizzled related protein 2 AB025922 Gli1, GLI-Kruppel family member 1 AF089721 Smo, Smoothened X99104 Gli2, GLI-Kruppel family member 2 AF009414 SOX11, SRY-box containing gene 11 U61362 Grg1, groucho-related gene 1, Tle1, transducin-like enhancer of split 1 U85614 SRG3, Smarcc1, SWI/SNF related, matrix associated, action dependent regulator of chromatin, subfamily, member 1 M97506 Hen1, helix-loop-helix protein AI837838 Tmeff1, Transmembrane protein with EGF-like and two follistatin-like domains 1 U79748 Madh4, MAD homolog 4, for transcription regulation -
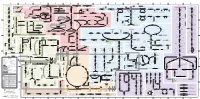
Fullsubwaymap221.Pdf
A B C D E F G H I J K L M Aldose reductase Sorbitol dehydrogenase Cholesterol synthesis Glycogen and galactose metabolism (branch point only) (debranching) glucose sorbitol fructose Aromatic amino acid metabolism Purine synthesis Pyrimidine synthesis glycogen (n+1) phenylacetate triiodothyronine (T3) dopaquinone melanin + + thyroxine (T4) (multiple steps) (many cycles) P 4' NADPH NADP NAD NADH acetyl-CoA x2 i Debranching enzyme 3' - α-1,4 bonds Aromatic amino acid 5-phosphoribosyl pyrophosphate (PRPP) HCO B 2' P 3 (branching) Glycogen 6 i (multiple steps) O H O (multiple steps) Tyrosinase O H O 1' 2 2 2 2 B 1 2 3 4 5 6 7 8 9 10 Pentose phosphate pathway Phenylalanine Tyrosine decarboxylase 6 Dopamine B Thiolase phosphorylase -5 -4 -3 -2 -1 0 1 2 3 4 Transaminase 6 glutamine 2 ATP (many cycles) Glycogen Glucose-6-phosphatase Dehydrogenase hydroxylase hydroxylase (DOPA decarboxylase) β-hydroxylase Methyltransferase 10 UDP 4:4 Transferase ATP glutathione (reduced) H O phenyllactate phenylpyruvate phenylalanine tyrosine dihydroxy- dopamine Glutamine PRPP CoA synthase Hexokinase or (in endoplasmic reticulum) 2 2 norepinephrine epinephrine glutamine Carbamoyl 9 1' phenylanine amidotransferase (GPAT) glutamate glycogen (n) Glutathione reductase Glutathione peroxidase + phosphate glycogen (n) -5 -4 -3 -2 -1 0 1 2 3 4 2' 3' 4' glucokinase 8 NADP NADPH glutamate α-ketoglutarate CO2 O H O Glycosyl (4:6) UDP-glucose ADP (L-DOPA) 2 2 synthetase II glutathione (oxidized) 2 H O α-ketoglutarate PPi glutamate acetoacetyl-CoA 7 1 Glucose -1,6-Glucosidase -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S.