Excel S2 SILAC Data HEK293T.Xlsx
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Datasheet Blank Template
SAN TA C RUZ BI OTEC HNOL OG Y, INC . DIP2A (L-16): sc-67555 BACKGROUND APPLICATIONS DIP2A (Disco-interacting protein 2 homolog A), also known as DIP2, is a 1,571 DIP2A (L-16) is recommended for detection of DIP2A of human origin by amino acid nuclear protein. It is one of three human homologs (DIP2A, DIP2B Western Blotting (starting dilution 1:200, dilution range 1:100-1:1000), and DIP2C) of the Drosophila dip2 (disconnected-interacting protein 2) protein. immunoprecipitation [1-2 µg per 100-500 µg of total protein (1 ml of cell In Drosophila , dip2 interacts with disco, a protein required for neuronal con - lysate)], immunofluorescence (starting dilution 1:50, dilution range 1:50- nections in the visual systems of larvae and adults. The closest vertebrate 1:500) and solid phase ELISA (starting dilution 1:30, dilution range 1:30- homologs to disco are the basonuclin genes. In mice, DIP2 homologs show 1:3000). restricted expression to the brain. This suggests that, similar to the function DIP2A (L-16) is also recommended for detection of DIP2A, also designated of Drosphila dip2, vertebrate DIP2 homologs may play a role in the develop - Disco-interacting protein 2 homolog A, in additional species, including ment of the nervous system. Expressed ubiquitously with highest expression canine. in the brain, DIP2A is thought to function in signaling throughout the central nervous system by providing positional clues for axon patterning and pathfind - Suitable for use as control antibody for DIP2A siRNA (h): sc-62212, DIP2A ing . Four isoforms of DIP2A exist due to alternative splicing events. -

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Total Synthesis and Chemoproteomics Connect Curcusone Diterpenes with Oncogenic Protein BRAT1
Total Synthesis and Chemoproteomics Connect Curcusone Diterpenes with Oncogenic Protein BRAT1 Chengsen Cui1†, Brendan G. Dwyer2†, Chang Liu1, Daniel Abegg2, Zhongjian Cai1, Dominic Hoch2, Xianglin Yin1, Nan Qiu2, Jieqing Liu3, Alexander Adibekian2*, Mingji Dai1* 1Department of Chemistry and Center for Cancer Research, Purdue University, West Lafayette, IN 47907, United States 2Department of Chemistry, The Scripps Research Institute, 130 Scripps Way, Jupiter, FL 33458, United States 3School of Medicine, Huaqiao University, Quanzhou 362021, P. R. China Correspondence and requests for materials should be addressed to M. D. (email: [email protected]) and A. A. ([email protected]) †Contributed equally. Abstract: Natural products are an indispensable source of lifesaving medicine, but natural product-based drug discovery often suffers from scarce natural supply and unknown mode of action. The study and development of anticancer curcusone diterpenes fall into such a dilemma. Meanwhile, many biologically- validated disease targets are considered “undruggable” due to the lack of enzymatic activity and/or predicted small molecule binding sites. The oncogenic BRCA1-associated ATM activator 1 (BRAT1) belongs to such an “undruggable” category. Here, we report our synthetic and chemoproteomics studies of the curcusone diterpenes that culminate in an efficient total synthesis and the identification of BRAT1 as a cellular target. We demonstrate for the first time that BRAT1 can be inhibited by a small molecule (curcusone D), resulting in impaired DNA damage response, reduced cancer cell migration, potentiated activity of the DNA damaging drug etoposide, and other phenotypes similar to BRAT1 knockdown. 1 Natural products have been valuable sources and inspirations of lifesaving drug molecules1. Their accumulated evolutionary wisdom together with their structural novelty and diversity makes them unparalleled for novel therapeutic development. -

SUPPLEMENTAL DATA Supplemental Materials And
SUPPLEMENTAL DATA Supplemental Materials and Methods Cells and Cell Culture Human breast carcinoma cell lines, MDA-MB-231 and MCF7, were purchased from American Type Tissue Culture Collection (ATCC). 231BoM-1833, 231BrM-2a, CN34, CN34-BoM2d, CN34-BrM2c and MCF7- BoM2d cell lines were kindly provided by Dr. Joan Massagué (Memorial Sloan-Kettering Cancer Center) (1-3). Luciferase-labeled cells were generated by infecting the lentivirus carrying the firefly luciferase gene. The immortalized mouse bone microvascular endothelial cell (mBMEC) was a generous gift from Dr. Isaiah J. Fidler (M.D. Anderson Cancer Center) (4). MCF10A and MCF10DCIS.com cells were purchased from ATCC and Asterand, respectively. MDA-MB-231, its variant cells, MCF7 and MCF-BoM2d cells were cultured in DMEM medium supplemented with 10% FBS and antibiotics. CN34 and its variant cells were cultured in Medium199 supplemented with 2.5% FBS, 10 µg/ml insulin, 0.5 µg/ml hydrocortisone, 20 ng/ml EGF, 100 ng/ml cholera toxin and antibiotics. MCF10DCIS.com cells were cultured in RPMI-1640 medium supplemented with 10% FBS and antibiotics. MCF10A cells were cultured in MEGM mammary epithelial cell growth medium (Lonza). mBMEC was maintained at 8% CO2 at 33 °C in DMEM with 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 1% non-essential amino acids and 1% vitamin mixture. Bone marrow stromal fibroblast cell lines HS5 and HS27A, and osteoblast cell line, hFOB1.19, were purchased from ATCC. Bone marrow derived human mesenchymal stem cells, BM-hMSC, were isolated for enrichment of plastic adherent cells from unprocessed bone marrow (Lonza) which was depleted of red blood cells. -

Identification and Characterization of TPRKB Dependency in TP53 Deficient Cancers
Identification and Characterization of TPRKB Dependency in TP53 Deficient Cancers. by Kelly Kennaley A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Molecular and Cellular Pathology) in the University of Michigan 2019 Doctoral Committee: Associate Professor Zaneta Nikolovska-Coleska, Co-Chair Adjunct Associate Professor Scott A. Tomlins, Co-Chair Associate Professor Eric R. Fearon Associate Professor Alexey I. Nesvizhskii Kelly R. Kennaley [email protected] ORCID iD: 0000-0003-2439-9020 © Kelly R. Kennaley 2019 Acknowledgements I have immeasurable gratitude for the unwavering support and guidance I received throughout my dissertation. First and foremost, I would like to thank my thesis advisor and mentor Dr. Scott Tomlins for entrusting me with a challenging, interesting, and impactful project. He taught me how to drive a project forward through set-backs, ask the important questions, and always consider the impact of my work. I’m truly appreciative for his commitment to ensuring that I would get the most from my graduate education. I am also grateful to the many members of the Tomlins lab that made it the supportive, collaborative, and educational environment that it was. I would like to give special thanks to those I’ve worked closely with on this project, particularly Dr. Moloy Goswami for his mentorship, Lei Lucy Wang, Dr. Sumin Han, and undergraduate students Bhavneet Singh, Travis Weiss, and Myles Barlow. I am also grateful for the support of my thesis committee, Dr. Eric Fearon, Dr. Alexey Nesvizhskii, and my co-mentor Dr. Zaneta Nikolovska-Coleska, who have offered guidance and critical evaluation since project inception. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
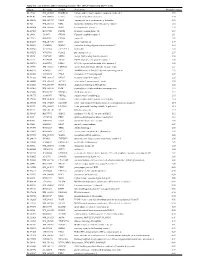
Supplementary Table 1
Table S1. List of Genes Differentially Expressed in YB-1 siRNA-Transfected MCF-7 Cells Unigene Accession Symbol Description Mean fold change Hs.17466 NM_004585 RARRES3 retinoic acid receptor responder (tazarotene induced) 3 3.57 Hs.64746 NM_004669 CLIC3 chloride intracellular channel 3 3.33 Hs.516155 NM_001747 CAPG capping protein (actin filament), gelsolin-like 2.88 Hs.926 NM_002463 MX2 myxovirus (influenza virus) resistance 2 (mouse) 2.72 Hs.643494 NM_005410 SEPP1 selenoprotein P, plasma, 1 2.70 Hs.267038 BC017500 POF1B premature ovarian failure 1B 2.67 Hs.20961 U63917 GPR30 G protein- coupled receptor 30 2532.53 Hs.199877 BE645967 CPNE4 copine IV 2.49 Hs.632177 NM_017458 MVP major vault protein 2.48 Hs.528836 AA005023 NOD27 nucleotide-binding oligomerization domains 27 2.43 Hs.360940 AL589866 dJ222E13.1 kraken-like 2.40 Hs.515575 AI743780 PLAC2 placenta-specific 2 2.37 Hs.25674 AI827820 MBD2 methyl-CpG binding domain protein 2 2.36 Hs.12229 AA149594 TIEG2 TGFB inducible early growth response 2 2.33 Hs.482730 AA053711 EDIL3 EGF-like repeats and discoidin I-like domains 3 2.32 Hs. 370771 NM_000389 CDKN1A cyclin- depen dent kinase in hibitor 1A (p 21, Cip 1) 2282.28 Hs.469175 AI341537 JFC1 NADPH oxidase-related, C2 domain-containing protein 2.28 Hs.514821 AF043341 CCL5 chemokine (C-C motif) ligand 5 2.27 Hs.591292 NM_023915 GPR87 G protein-coupled receptor 87 2.26 Hs.500483 NM_001613 ACTA2 actin, alpha 2, smooth muscle, aorta 2.24 Hs.632824 NM_006729 DIAPH2 diaphanous homolog 2 (Drosophila) 2.22 Hs.369430 NM_000919 PAM peptidylglycine -

Supplementary Materials
1 Supplementary Materials: Supplemental Figure 1. Gene expression profiles of kidneys in the Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice. (A) A heat map of microarray data show the genes that significantly changed up to 2 fold compared between Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice (N=4 mice per group; p<0.05). Data show in log2 (sample/wild-type). 2 Supplemental Figure 2. Sting signaling is essential for immuno-phenotypes of the Fcgr2b-/-lupus mice. (A-C) Flow cytometry analysis of splenocytes isolated from wild-type, Fcgr2b-/- and Fcgr2b-/-. Stinggt/gt mice at the age of 6-7 months (N= 13-14 per group). Data shown in the percentage of (A) CD4+ ICOS+ cells, (B) B220+ I-Ab+ cells and (C) CD138+ cells. Data show as mean ± SEM (*p < 0.05, **p<0.01 and ***p<0.001). 3 Supplemental Figure 3. Phenotypes of Sting activated dendritic cells. (A) Representative of western blot analysis from immunoprecipitation with Sting of Fcgr2b-/- mice (N= 4). The band was shown in STING protein of activated BMDC with DMXAA at 0, 3 and 6 hr. and phosphorylation of STING at Ser357. (B) Mass spectra of phosphorylation of STING at Ser357 of activated BMDC from Fcgr2b-/- mice after stimulated with DMXAA for 3 hour and followed by immunoprecipitation with STING. (C) Sting-activated BMDC were co-cultured with LYN inhibitor PP2 and analyzed by flow cytometry, which showed the mean fluorescence intensity (MFI) of IAb expressing DC (N = 3 mice per group). 4 Supplemental Table 1. Lists of up and down of regulated proteins Accession No. -

The Interactome of KRAB Zinc Finger Proteins Reveals the Evolutionary History of Their Functional Diversification
Resource The interactome of KRAB zinc finger proteins reveals the evolutionary history of their functional diversification Pierre-Yves Helleboid1,†, Moritz Heusel2,†, Julien Duc1, Cécile Piot1, Christian W Thorball1, Andrea Coluccio1, Julien Pontis1, Michaël Imbeault1, Priscilla Turelli1, Ruedi Aebersold2,3,* & Didier Trono1,** Abstract years ago (MYA) (Imbeault et al, 2017). Their products harbor an N-terminal KRAB (Kru¨ppel-associated box) domain related to that of Krüppel-associated box (KRAB)-containing zinc finger proteins Meisetz (a.k.a. PRDM9), a protein that originated prior to the diver- (KZFPs) are encoded in the hundreds by the genomes of higher gence of chordates and echinoderms, and a C-terminal array of zinc vertebrates, and many act with the heterochromatin-inducing fingers (ZNF) with sequence-specific DNA-binding potential (Urru- KAP1 as repressors of transposable elements (TEs) during early tia, 2003; Birtle & Ponting, 2006; Imbeault et al, 2017). KZFP genes embryogenesis. Yet, their widespread expression in adult tissues multiplied by gene and segment duplication to count today more and enrichment at other genetic loci indicate additional roles. than 350 and 700 representatives in the human and mouse Here, we characterized the protein interactome of 101 of the ~350 genomes, respectively (Urrutia, 2003; Kauzlaric et al, 2017). A human KZFPs. Consistent with their targeting of TEs, most KZFPs majority of human KZFPs including all primate-restricted family conserved up to placental mammals essentially recruit KAP1 and members target sequences derived from TEs, that is, DNA trans- associated effectors. In contrast, a subset of more ancient KZFPs posons, ERVs (endogenous retroviruses), LINEs, SINEs (long and rather interacts with factors related to functions such as genome short interspersed nuclear elements, respectively), or SVAs (SINE- architecture or RNA processing. -

A Bootstrap-Based Regression Method for Comprehensive Discovery of Differential Gene Expressions: an Application to the Osteoporosis Study
A bootstrap-based regression method for comprehensive discovery of differential gene expressions: an application to the osteoporosis study Yan Lu1,2, Yao-Zhong Liu3, Peng–Yuan Liu2, Volodymyr Dvornyk4, and Hong–Wen Deng1,3 1. College of Life Sciences and Bioengineering, Beijing Jiaotong University, Beijing 100044, P. R. China 2. Department of Physiology and the Cancer Center, Medical College of Wisconsin, Milwaukee, WI 53226, USA 3. Department of Biostatistics and Bioinformatics & Center for Bioinformatics and Genomics, Tulane University School of Public Health, New Orleans, LA 70112, USA 4. School of Biological Sciences, University of Hong Kong, Pokfulam Rd., Pokfulam, Hong Kong, P.R. China Running title: Bootstrap-based regression method for microarray analysis Key words: microarray, bootstrap, regression, osteoporosis Corresponding author: Hong–Wen Deng, Ph. D. Department of Biostatistics and Bioinformatics & Center for Bioinformatics and Genomics, Tulane University School of Public Health, 1440 Canal Street, Suite 2001, New Orleans, LA 70112 Tel: 504-988-1310 Email: [email protected] 1 Abstract A common purpose of microarray experiments is to study the variation in gene expression across the categories of an experimental factor such as tissue types and drug treatments. However, it is not uncommon that the studied experimental factor is a quantitative variable rather than categorical variable. Loss of information would occur by comparing gene-expression levels between groups that are factitiously defined according to the quantitative threshold values of an experimental factor. Additionally, lack of control for some sensitive clinical factors may bring serious false positive or negative findings. In the present study, we described a bootstrap-based regression method for analyzing gene expression data from the non-categorical microarray experiments. -

Noelia Díaz Blanco
Effects of environmental factors on the gonadal transcriptome of European sea bass (Dicentrarchus labrax), juvenile growth and sex ratios Noelia Díaz Blanco Ph.D. thesis 2014 Submitted in partial fulfillment of the requirements for the Ph.D. degree from the Universitat Pompeu Fabra (UPF). This work has been carried out at the Group of Biology of Reproduction (GBR), at the Department of Renewable Marine Resources of the Institute of Marine Sciences (ICM-CSIC). Thesis supervisor: Dr. Francesc Piferrer Professor d’Investigació Institut de Ciències del Mar (ICM-CSIC) i ii A mis padres A Xavi iii iv Acknowledgements This thesis has been made possible by the support of many people who in one way or another, many times unknowingly, gave me the strength to overcome this "long and winding road". First of all, I would like to thank my supervisor, Dr. Francesc Piferrer, for his patience, guidance and wise advice throughout all this Ph.D. experience. But above all, for the trust he placed on me almost seven years ago when he offered me the opportunity to be part of his team. Thanks also for teaching me how to question always everything, for sharing with me your enthusiasm for science and for giving me the opportunity of learning from you by participating in many projects, collaborations and scientific meetings. I am also thankful to my colleagues (former and present Group of Biology of Reproduction members) for your support and encouragement throughout this journey. To the “exGBRs”, thanks for helping me with my first steps into this world. Working as an undergrad with you Dr.