Understanding the Transcriptional Regulation of Secondary Cell Wall Biosynthesis in the Model Grass Brachypodium Distachyon
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Eucalyptus Gunnii Subsp. Divaricata (Mcaulay & Brett) B.M
Listing StatementEucalyptus for Eucalyptus gunnii gunnii subsp. subsp. divaricata (miena divaricata cider gum) miena cider gum T A S M A N I A N T H R E A T E N E D S P E C I E S L I S T I N G S T A T E M E N T Image by B. Potts Scientific name: Eucalyptus gunnii subsp. divaricata (McAulay & Brett) B.M. Potts, Pap. Proc. R. Soc. Tasm . 135: 57 (2001) Common name: miena cider gum (Wapstra et al. 2005) Group: vascular plant, dicotyledon, family Myrtaceae Status: Threatened Species Protection Act 1995 : endangered Environment Protection and Biodiversity Conservation Act 1999 : Endangered Distribution Endemic status: Endemic to Tasmania Tasmanian NRM Region: South Figure 1. Distribution of specimens attributed to Plate 1 . Eucalyptus gunnii subsp. divaricata Eucalyptus gunnii subsp. divaricata . The northern and (Image by J. Calder) western most records require verification. 1 Threatened Species Section – Department of Primary Industries, Parks, Water and Environment Listing Statement for Eucalyptus gunnii subsp. divaricata (miena cider gum) IDENTIFICATION AND ECOLOGY in drought conditions or in stands disturbed by Eucalyptus gunnii subsp. divaricata is a small to stock grazing. While Eucalyptus gunnii subsp. medium sized tree in the Myrtaceae family divaricata is highly frost resistant, it is the first (Plate 1). It is endemic to Tasmania’s Central eucalypt in the area to display symptoms Plateau where it mostly grows on the edges of following drought, leading to the death of frost hollows (Potts et al. 2001). Eucalyptus gunnii mature trees in relatively large patches since the subsp. -

Genetic Diversity and Adaptation in Eucalyptus Pauciflora
Genetic diversity and adaptation in Eucalyptus pauciflora Archana Gauli (M.Sc.) A thesis submitted in fulfilment of the requirements for the Degree of Doctor of Philosophy School of Biological Sciences, University of Tasmania June, 2014 Declarations This thesis contains no material which has been accepted for a degree or diploma by the University or any other institution, except by way of background information and duly acknowledged in the thesis, and to the best of the my knowledge and belief no material previously published or written by another person except where due acknowledgement is made in the text of the thesis, nor does the thesis contain any material that infringes copyright. Archana Gauli Date Authority of access This thesis may be made available for loan and limited copying and communication in accordance with the Copyright Act 1968. Archana Gauli Date Statement regarding published work contained in thesis The publishers of the paper comprising Chapter 2 and Chapter 3 hold the copyright for that content, and access to the material should be sought from the respective journals. The remaining non-published content of the thesis may be made available for loan and limited copying and communication in accordance with the Copyright Act 1968. Archana Gauli Date i Statement of publication Chapter 2 has been published as: Gauli A, Vaillancourt RE, Steane DA, Bailey TG, Potts BM (2014) The effect of forest fragmentation and altitude on the mating system of Eucalyptus pauciflora (Myrtaceae). Australian Journal of Botany 61, 622-632. Chapter 3 has been accepted for publication as: Gauli A, Steane DA, Vaillancourt RE, Potts BM (in press) Molecular genetic diversity and population structure in Eucalyptus pauciflora subsp. -

Eucalyptus That Are Ideal for British Gardens
Barks, shoots and leaves Grafton Nursery grows hardy forms of Antipodean Eucalyptus that are ideal for British gardens WORDS JOHN HOYLAND PHOTOGRAPHS JASON INGRAM Gower Hilary Collins checks the growth on a selection of the hardy eucalyptus grown at Grafton Nursery. Nearby assistant grower Adam Barnes keeps the more mature trees in check with some careful pruning. 68 FOR CUTTING 1 Eucalyptus ‘Shannon Blue’ VERY FEW TREES ARE AS FASCINATING A hybrid bred for the floristry trade, this vigorous plant has stems of rounded, glaucous, silver AS EUCALYPTS: THEY ARE HIGHLY leaves. It can be grown as a hedge and retains EVOLVED AND EXTREMELY VARIED its colour through the winter. Unpruned height 10m. RHS H5†. 2 Eucalyptus nicholii An airy tree with narrow, almost feathery, sage- green leaves. Its common name – narrow-leaved black peppermint – references its strongly aromatic foliage. Unpruned height 12m. RHS H5. 3 Eucalyptus pulverulenta ‘Baby Blue’ ardeners can be blinkered, disregarding a whole A compact, bushy cultivar that can be grown in a pot. The silvery-grey stems are long-lasting genus because of the faults of a few members. when cut for flower arrangements. Unpruned Eucalyptus has suffered this fate, largely because of height 5m. RHS H5. the thuggish behaviour of Eucalyptus gunnii, the most widely available species, which will grow to huge 4 Eucalyptus gunnii proportions in a very short time. “There are far better The most widely grown species in the UK. It has peeling cream and brown bark, rounded, species,” says grower Hilary Collins. “Beautiful glaucous-blue juvenile leaves and elliptic or eucalypts for small terraces or large pots, those for sickle-shaped, grey-green adult leaves. -

ADAPTATION of FORESTS and PEOPLE to CLIMATE Change – a Global Assessment Report
International Union of Forest Research Organizations Union Internationale des Instituts de Recherches Forestières Internationaler Verband Forstlicher Forschungsanstalten Unión Internacional de Organizaciones de Investigación Forestal IUFRO World Series Vol. 22 ADAPTATION OF FORESTS AND PEOPLE TO CLIMATE CHANGE – A Global Assessment Report Prepared by the Global Forest Expert Panel on Adaptation of Forests to Climate Change Editors: Risto Seppälä, Panel Chair Alexander Buck, GFEP Coordinator Pia Katila, Content Editor This publication has received funding from the Ministry for Foreign Affairs of Finland, the Swedish International Development Cooperation Agency, the United Kingdom´s Department for International Development, the German Federal Ministry for Economic Cooperation and Development, the Swiss Agency for Development and Cooperation, and the United States Forest Service. The views expressed within this publication do not necessarily reflect official policy of the governments represented by these institutions. Publisher: International Union of Forest Research Organizations (IUFRO) Recommended catalogue entry: Risto Seppälä, Alexander Buck and Pia Katila. (eds.). 2009. Adaptation of Forests and People to Climate Change. A Global Assessment Report. IUFRO World Series Volume 22. Helsinki. 224 p. ISBN 978-3-901347-80-1 ISSN 1016-3263 Published by: International Union of Forest Research Organizations (IUFRO) Available from: IUFRO Headquarters Secretariat c/o Mariabrunn (BFW) Hauptstrasse 7 1140 Vienna Austria Tel: + 43-1-8770151 Fax: + 43-1-8770151-50 E-mail: [email protected] Web site: www.iufro.org/ Cover photographs: Matti Nummelin, John Parrotta and Erkki Oksanen Printed in Finland by Esa-Print Oy, Tampere, 2009 Preface his book is the first product of the Collabora- written so that they can be read independently from Ttive Partnership on Forests’ Global Forest Expert each other. -

The Natural Distribution of Eucalyptus Species in Tasmania
The natural distribution of Eucalyptus species in Tasmania K.J. Williams and B.M. Potts Cooperative Research Centre for Temperate Hardwood Forestry, Department of Plant Science, University of Tasmania, GPO Box 252–55, Hobart 7001 email: [email protected]./[email protected] Abstract dispersed (E. cordata) or disjunct (E. archeri) occurrences. Most species that are rare in A summary is provided of the natural geographic Tasmania are endemics, with the exception of distributions of the 29 Tasmanian Eucalyptus E. perriniana and E. aff. radiata, although species. The work is based on over 60 000 the taxonomic status of the latter requires observations from numerous data sources. A map investigation. Unresolved issues relating to the on a 10 km x 10 km grid-cell scale is presented for natural distribution and taxonomic affinities of each species and is accompanied by graphs of the the Tasmanian eucalypt species are summarised. altitudinal range and flowering times, as well as descriptive notes on distribution and ecology, supplemented with a list of key references. The Introduction geographic pattern of species richness is examined at generic, subgeneric and series levels. Total In Tasmania and the Bass Strait islands, species richness is greater in the drier, eastern 29 native eucalypt species (one of which has regions compared to the wet, western regions of two subspecies) are recognised by Buchanan Tasmania, with highest concentrations of species (1995), from two informal subgenera, occurring mainly in the central east coast and Monocalyptus and Symphyomyrtus (Pryor and south-eastern regions. Monocalyptus species Johnson 1971). -

Critical Revision of the Genus Eucalyptus Volume 3: Parts 21-30
Critical revision of the genus eucalyptus Volume 3: Parts 21-30 Maiden, J. H. (Joseph Henry) (1859-1925) University of Sydney Library Sydney 2002 http://setis.library.usyd.edu.au/oztexts © University of Sydney Library. The texts and images are not to be used for commercial purposes without permission Source Text: Prepared from the print edition of Parts 21-30 Critical revision of the genus eucalyptus, published by William Applegate Gullick Sydney 1917. 223pp. All quotation marks are retained as data. First Published: 1917 583.42 Australian Etext Collections at botany prose nonfiction 1910-1939 Critical revision of the genus eucalyptus volume 3 (Government Botanist of New South Wales and Director of the Botanic Gardens, Sydney) “Ages are spent in collecting materials, ages more in separating and combining them. Even when a system has been formed, there is still something to add, to alter, or to reject. Every generation enjoys the use of a vast hoard bequeathed to it by antiquity, and transmits that hoard, augmented by fresh acquisitions, to future ages. In these pursuits, therefore, the first speculators lie under great disadvantages, and, even when they fail, are entitled to praise.” Macaulay's “Essay on Milton” Sydney William Applegate Gullick, Government Printer 1917 Part 21 CXIII. E. cinerea F.v.M. In Bentham's Flora Australiensis iii, 239 (1866). FOLLOWING is the original description:— A moderate-sized tree, with a whitish-brown persistent bark, somewhat fibrous, the foliage more or less glaucous or mealy white. Leaves opposite, sessile, cordate ovate or ovate-lanceolate, obtuse or acute, mostly 2 to 4 inches long (or narrow lanceolate, which are alternate and much longer.—J.H.M.). -

The Vegetation Communities Dry Eucalypt Forest and Woodland
Edition 2 From Forest to Fjaeldmark The Vegetation Communities Dry eucalypt forest and woodland Eucalyptus amygdalina Edition 2 From Forest to Fjaeldmark 1 Dry eucalypt forest and woodland Community (Code) Page Eucalyptus amygdalina coastal forest and woodland (DAC) 11 Eucalyptus amygdalina forest and woodland on dolerite (DAD) 13 Eucalyptus amygdalina forest and woodland on sandstone (DAS) 15 Eucalyptus amygdalina forest on mudstone (DAM) 17 Eucalyptus amygdalina inland forest and woodland on Cainozoic deposits (DAZ) 19 Eucalyptus amygdalina–Eucalyptus obliqua damp sclerophyll forest (DSC) 22 Eucalyptus barberi forest and woodland (DBA) 24 Eucalyptus coccifera forest and woodland (DCO) 25 Eucalyptus cordata forest (DCR) 27 Eucalyptus dalrympleana–Eucalyptus pauciflora forest and woodland (DDP) 29 Eucalyptus delegatensis dry forest and woodland (DDE) 31 Eucalyptus globulus dry forest and woodland (DGL) 33 Eucalyptus gunnii woodland (DGW) 35 Eucalyptus morrisbyi forest and woodland (DMO) 37 Eucalyptus nitida dry forest and woodland (DNI) 39 Eucalyptus nitida Furneaux forest (DNF) 41 Eucalyptus obliqua dry forest (DOB) 43 Eucalyptus ovata forest and woodland (DOV) 45 Eucalyptus ovata heathy woodland (DOW) 48 Eucalyptus pauciflora forest and woodland not on dolerite (DPO) 50 Eucalyptus pauciflora forest and woodland on dolerite (DPD) 52 Eucalyptus perriniana forest and woodland (DPE) 54 Eucalyptus pulchella forest and woodland (DPU) 56 Eucalyptus risdonii forest and woodland (DRI) 58 Eucalyptus rodwayi forest and woodland (DRO) 60 Eucalyptus -

ABSTRACT the First Through Fifth Instars of the Gypsy Moth Were Tested for Development to Adults on 326 Species of Dicotyledonous Plants in Laboratory Feeding Trials
LABORATORY FEEDING TESTS ON THE DEVELOPMENT OF GYPSY MOTH LARVAE WITH REFERENCE TO PLANT TAXA AND ALLELOCHEMICALS JEFFREY C. MILLER and PAUL E. HANSON DEPARTMENT OF ENTOMOLOGY, OREGON STATE UNIVERSITY, CORVALLIS, OREGON 97331 ABSTRACT The first through fifth instars of the gypsy moth were tested for development to adults on 326 species of dicotyledonous plants in laboratory feeding trials. Among accepted plants, differences in suitability were documented by measuring female pupal weights. The majority of accepted plants belong to the subclasses Dilleniidae, Hamamelidae, and Rosidae. Species of oak, maple, alder, madrone, eucalyptus, poplar, and sumac were highly suitable. Plants belonging to the Asteridae, Caryophyllidae, and Magnoliidae were mostly rejected. Foliage type, new or old, and instar influenced host plant suitability. Larvae of various instars were able to pupate after feeding on foliage of 147 plant species. Of these, 1.01 were accepted by first instars. Larvae from the first through fifth instar failed to molt on foliage of 151 species. Minor feeding occurred on 67 of these species. In general, larvae accepted new foliage on evergreen species more readily than old foliage. The results of these trials were combined with results from three previous studies to provide data on feeding responses of gypsy moth larvae on a total of 658 species, 286 genera, and 106 families of dicots. Allelochemic compositions of these plants were tabulated from available literature and compared with acceptance or rejection by gypsy moth. Plants accepted by gypsy moth generally contain tannins, but lack alkaloids, iridoid monoterpenes, sesquiterpenoids, diterpenoids, and glucosinolates. 2 PREFACE This research was funded through grants from USDA Forest Service cooperative agreement no. -

Eucalyptus for Cut Foliage Factsheet 2/20
Eucalyptus for Cut Foliage Factsheet 2/20 INTRODUCTION SPECIES Eucalyptus originates from Australia and consists of The main requirements of a species for cutting are: over 500 different species many of which grow well • Attractive foliage colour and form. in the South of Ireland. There is demand for Eucalypts • Ability to coppice with rapid annual branching. with attractive grey, blue-grey and green foliage all • At least some resistance to low winter temperature. year round and particularly during peak autumn, winter and spring months. The stems are used as ‘fillers’ in Markets vary in their preference to particular species so mixed flower bouquets supplying high street florists, an it is important to thoroughly research the market before export market to specialist flower bouquet processing establishing a plantation. companies in the UK and Holland who in turn supply the The following species have performed well in Ireland. major supermarkets and other retail outlets including an expanding mail order market. The first commercial Eucalyptus cinerea (Silver Dollar) – probably the most plantings were made in Ireland in the early 90’s and the popular grey species. Small leaves and purple young current area stands at approximately 100 ha. stems. Immature leaves pink. Very hardy, coppices well. Medium to high yields. Eucalyptus coccifera (Tasmanian Snow Gum) – juvenile leaves smell of peppermint. Glistening young shoots, willow like blue adult leaves. Very hardy and tolerates exposure. Eucalyptus glaucescens (Tingiringi Gum) – small rounded leaves blue/white with a ‘fruity’ aroma. Young adult leaves glaucous or pink. Coppices well. Medium yields. Eucalyptus gunnii (The Cider Gum) – leaves rounded, surface powdery blue-grey. -
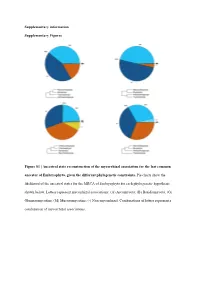
Ancestral State Reconstruction of the Mycorrhizal Association for the Last Common Ancestor of Embryophyta, Given the Different Phylogenetic Constraints
Supplementary information Supplementary Figures Figure S1 | Ancestral state reconstruction of the mycorrhizal association for the last common ancestor of Embryophyta, given the different phylogenetic constraints. Pie charts show the likelihood of the ancestral states for the MRCA of Embryophyta for each phylogenetic hypothesis shown below. Letters represent mycorrhizal associations: (A) Ascomycota; (B) Basidiomycota; (G) Glomeromycotina; (M) Mucoromycotina; (-) Non-mycorrhizal. Combinations of letters represent a combination of mycorrhizal associations. Austrocedrus chilensis Chamaecyparis obtusa Sequoiadendron giganteum Prumnopitys taxifolia Prumnopitys Prumnopitys montana Prumnopitys Prumnopitys ferruginea Prumnopitys Araucaria angustifolia Araucaria Dacrycarpus dacrydioides Dacrycarpus Taxus baccata Podocarpus oleifolius Podocarpus Afrocarpus falcatus Afrocarpus Ephedra fragilis Nymphaea alba Nymphaea Gnetum gnemon Abies alba Abies balsamea Austrobaileya scandens Austrobaileya Abies nordmanniana Thalictrum minus Thalictrum Abies homolepis Caltha palustris Caltha Abies magnifica ia repens Ranunculus Abies religiosa Ranunculus montanus Ranunculus Clematis vitalba Clematis Keteleeria davidiana Anemone patens Anemone Tsuga canadensis Vitis vinifera Vitis Tsuga mertensiana Saxifraga oppositifolia Saxifraga Larix decidua Hypericum maculatum Hypericum Larix gmelinii Phyllanthus calycinus Phyllanthus Larix kaempferi Hieronyma oblonga Hieronyma Pseudotsuga menziesii Salix reinii Salix Picea abies Salix polaris Salix Picea crassifolia Salix herbacea -

Climate Adaptation and Provenance Choice for Revegetation: Insights from Eucalyptus
Climate adaptation and provenance choice for revegetation: Insights from Eucalyptus Brad Potts Peter Harrison, Tanya Bailey, Achana Gauli, Dorothy Steane, René Vaillancourt, Paul Tilyard, Neil Davidson 1. Eucalypt populations exhibit a close adaptive response to their environment • local-scale e.g. altitudinal or edaphic • broad-scale e.g. macro-climate LD Pryor WD Jackson Pryor LD (1957) Proc. Linn. Soc. NSW 81: 299-305 Barber & Jackson (1957) Nature 179: 1267-1269 Numerous lines of evidence for climate adaptation 1. Correlative - evidence of divergent selection e.g. divergence in functional trait (QST) exceeds neutral molecular expectations (drift) (FST) - plus correlated with spatial variation in climate - plus consistency with functional expectations 2. Direct (selection experiments) - laboratory/glasshouse - field trials: - in situ (e.g. reciprocal plantings in the wild) - ex situ (e.g. multi-site tree breeding<3m trials) tall 40 - 70m tall 3 Range-wide seed collections and testing in breeding trials: the case of E. globulus Sampling localities CSIRO 1987/88 base population seed collection - 600+ open-pollinated families - 46 localities I Trials designed with II randomization at the family level (e.g. III randomized incomplete block designs, 2 tree IV plots, 5 reps) V Dutkowski and Potts 1999 Lopez et al. 2001 Seed lots from the 87/88 CSIRO collection of E. globulus have been tested globally 76 International trials 26 Australian trials Spain China Portugal Columbia Chile Ethiopia Tasmania Western Victoria Australia South Australia Potts et al. 2004 Home-site climate predicts susceptibility to drought and growth on dry sites Drought tolerance Tree growth (dbh) Strzelecki Ranges high Relative performance Relative Furneaux Group 0 Prevailing King Island Tolerant winds 79,730 trees low Tolerant Tolerant dry wet 22,245 trees Rainfall in warmest quarter at site Intolerant Costa e Silva et al. -

Eucalyptus Genus: a Review
Gagan Shahet al. / Journal of Pharmacy Research 2016,10(10),609-617 Review Article Available online through ISSN: 0974-6943 http://jprsolutions.info Eucalyptus Genus: A Review Gagan Shah*1, Jaideep Bajaj2, Varinder Soni3, R.K Dhawan2 *Deparment of Pharmacognosy, Khalsa College of Pharmacy, Amritsar, Punjab, India 2Department of Pharmacology, Khalsa College of Pharmacy, Amritsar Punjab, India 3Department of Pharmaceutical Analysis, Khalsa College of Pharmacy, Amritsar Punjab, India Received on:25-07-2016; Revised on: 11-08-2016; Accepted on: 04-09-2016 ABSTRACT Eucalyptus is a diverse genus of flowering trees (and a few shrubs) in the myrtle family, Myrtaceae. This review article is presented to compile all the updated information on its phytochemical and pharmacological Activities of Eucalyptus species which were performed by widely different methods. Literature indicates various Eucalyptus Species possesses analgesic, antifungal, antiinflamatory, antibacterial, antidiabetic, antioxidative properties. Literature also indicates various other effect of Eucalyptus species such as Antiviral, Antitumour, antihistaminic, anticancer cytochrome p450 inhibitor and hepatoprotective effect have also been reported by many Researcher. The present review articles critically discusses some Eucalyptus Species and about their Chemical constituents and Biological activities. This review indicate that Eucalyptus species have potential therapeutic effects. KEY WORDS: Eucalyptus, Phytochemical, Pharmacological activities. INTRODUCTION MAJOR SPECIES A large genus of evergreen aromatic tress, rarely shrubs (mallees), There are over 500 species of Eucalyptus. The major ones are enlisted indeginous to Australia, Tasmania, New Guinea and the neighbouring below. Major Species of Eucalyptus. Major Species of Eucalyptus. islands, where they constitute a large portion of the forest vegetation, Eucalyptus amygdalina Eucalyptus microthecaamygdalina giving it a characteristic appearance.