Little Brown Bat (Myotis Lucifugus) BAC Library
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The World at the Time of Messel: Conference Volume
T. Lehmann & S.F.K. Schaal (eds) The World at the Time of Messel - Conference Volume Time at the The World The World at the Time of Messel: Puzzles in Palaeobiology, Palaeoenvironment and the History of Early Primates 22nd International Senckenberg Conference 2011 Frankfurt am Main, 15th - 19th November 2011 ISBN 978-3-929907-86-5 Conference Volume SENCKENBERG Gesellschaft für Naturforschung THOMAS LEHMANN & STEPHAN F.K. SCHAAL (eds) The World at the Time of Messel: Puzzles in Palaeobiology, Palaeoenvironment, and the History of Early Primates 22nd International Senckenberg Conference Frankfurt am Main, 15th – 19th November 2011 Conference Volume Senckenberg Gesellschaft für Naturforschung IMPRINT The World at the Time of Messel: Puzzles in Palaeobiology, Palaeoenvironment, and the History of Early Primates 22nd International Senckenberg Conference 15th – 19th November 2011, Frankfurt am Main, Germany Conference Volume Publisher PROF. DR. DR. H.C. VOLKER MOSBRUGGER Senckenberg Gesellschaft für Naturforschung Senckenberganlage 25, 60325 Frankfurt am Main, Germany Editors DR. THOMAS LEHMANN & DR. STEPHAN F.K. SCHAAL Senckenberg Research Institute and Natural History Museum Frankfurt Senckenberganlage 25, 60325 Frankfurt am Main, Germany [email protected]; [email protected] Language editors JOSEPH E.B. HOGAN & DR. KRISTER T. SMITH Layout JULIANE EBERHARDT & ANIKA VOGEL Cover Illustration EVELINE JUNQUEIRA Print Rhein-Main-Geschäftsdrucke, Hofheim-Wallau, Germany Citation LEHMANN, T. & SCHAAL, S.F.K. (eds) (2011). The World at the Time of Messel: Puzzles in Palaeobiology, Palaeoenvironment, and the History of Early Primates. 22nd International Senckenberg Conference. 15th – 19th November 2011, Frankfurt am Main. Conference Volume. Senckenberg Gesellschaft für Naturforschung, Frankfurt am Main. pp. 203. -
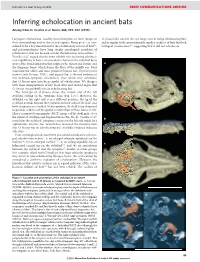
Inferring Echolocation in Ancient Bats Arising From: N
NATURE | Vol 466 | 19 August 2010 BRIEF COMMUNICATIONS ARISING Inferring echolocation in ancient bats Arising from: N. Veselka et al. Nature 463, 939–942 (2010) Laryngeal echolocation, used by most living bats to form images of O. finneyi falls outside the size range seen in living echolocating bats their surroundings and to detect and capture flying prey1,2, is con- and is similar to the proportionally smaller cochleae of bats that lack sidered to be a key innovation for the evolutionary success of bats2,3, laryngeal echolocation4,8, suggesting that it did not echolocate. and palaeontologists have long sought osteological correlates of echolocation that can be used to infer the behaviour of fossil bats4–7. Veselka et al.8 argued that the most reliable trait indicating echoloca- tion capabilities in bats is an articulation between the stylohyal bone (part of the hyoid apparatus that supports the throat and larynx) and a the tympanic bone, which forms the floor of the middle ear. They examined the oldest and most primitive known bat, Onychonycteris finneyi (early Eocene, USA4), and argued that it showed evidence of this stylohyal–tympanic articulation, from which they concluded that O. finneyi may have been capable of echolocation. We disagree with their interpretation of key fossil data and instead argue that O. finneyi was probably not an echolocating bat. The holotype of O. finneyi shows the cranial end of the left stylohyal resting on the tympanic bone (Fig. 1c–e). However, the stylohyal on the right side is in a different position, the tip of the stylohyal extends beyond the tympanic on both sides of the skull, and both tympanics are crushed. -

103 the EVOLUTION of BATSI Leigh Van Valen Department Of
103 THE EVOLUTIONOF BATSI Leigh Van Valen Department of Biology University of Chicago 1103 East 57th Street Chicago, Illinois 60637 Received August 30, 1978; June 8, L979 Abstract: I devel-op the first explicitly Justified phylogeny of the known farnil-les of bats, usj.ng all available characters. This phylogeny permits the adaptive evolution of the order to be outlined. Two main clades emerge within the Microchiroptera and are cal-led new infraorders, Vespertilionia and Phyllostomatia. In each infraorder there is a series of grades of progressively stronger fi-ight, and there is a radiation of diet within the Phyllostomatia. Parallel evolution is extensive. Most grades stlll exist, presumably by a poorly understood partitioning of the resource space. The Megachiroptera nay have originated in the l-ate Ollgocene or early Miocene from surviving mernbers of the Eochiroptera (new suborder). The Kerivoul,ldae, Myzopodidae, Thyropteridae, and Furlpterldae are placed in the Natalidae, and the lcaronycterididae in the PaLaeochiropterygidae. The features of an ancestral bat are predicted. Bat origins are poorly known but may be found in Paleocene members of the Adapisoricidae. ,r** Bats constitute the second largest order of marnmalsand have a readily decipherable adaptive history, at least compared with the Rodentia and Insectivora. Nevertheless, there is no treatment of this adaptive hlstory nor even a general phyl-ogeny, which must form its foundation. I noticed this l-ack when revising a course on the paleobiology of mauunalsand undertook to remedy it. Although I sttll lack much familiarity with bats, the resul-t may be of more general interest. ORIGIN OF BATS One may hypothesize ttrat bats dld originate, but it is harder to go beyond this. -

Messel Pit – Wikipedia Germany
03/08/2018 Messel pit - Wikipedia Coordinates: 49°55′03″N 8°45′24″E Messel pit The Messel Pit (German: Grube Messel) is a disused quarry near the Messel Pit Fossil Site village of Messel, (Landkreis Darmstadt-Dieburg, Hesse) about 35 km (22 mi) southeast of Frankfurt am Main, Germany. Bituminous shale UNESCO World Heritage site was mined there. Because of its abundance of fossils, it has significant geological and scientific importance. After almost becoming a landfill, strong local resistance eventually stopped these plans and the Messel Pit was declared a UNESCO World Heritage site on 9 December 1995. Significant scientific discoveries are still being made and the site has increasingly become a tourist site as well. Contents Location Darmstadt-Dieburg, History Hesse, Germany Depositional characteristics Criteria Natural: (viii) Volcanic gas releases Reference 720bis (http://whc.unesco. Fossils org/en/list/720bis) Mammals Inscription 1995 (19th Session) Birds Reptiles Extensions 2010 Fish Area 42 ha (4,500,000 sq ft) Insects Plants Buffer zone 22.5 ha (2,420,000 sq ft) Access Coordinates 49°55′03″N 8°45′24″E See also References External links History Brown coal and later oil shale was actively mined from 1859. The pit first became known for its wealth of fossils around 1900, but serious scientific excavation only started around the 1970s, when falling oil prices made the quarry uneconomical. Commercial oil shale mining ceased in 1971 and a cement factory built in the quarry failed the following year. The land was slotted for use as a landfill, but the plans came to nought and the Hessian state bought the site in 1991 to secure scientific access. -

To Scream Or to Listen? Prey Detection and Discrimination in Animal-Eating Bats
96 P.L. Jones et al. Fig. 4.1 Current phylogeny for bats (Jones and Teeling 2006 ). To the right of each family name, a Substrate Gleaner icon indicates that, in our opinion, this family is characterized by bat species that rely primarily on a gleaning strategy; all or most of which also take some prey by aerial hawk- ing. An Aerial Hawker icon indicates that the family consists of species most of which primarily use a hawking strategy but includes behaviorally fl exible species. Crasoenycteridae comprises a single behaviorally fl exible species. A Frugivore/Nectivore icon indicates a family comprised solely or partially of frugivorous and nectivorous species Each scenario, however, still suggests the same general story (Figure 4.1 ). That is, early laryngeal echolocating bats used powered fl ight, hunted animals, and took them in the air. The latter supposition is supported by the fact that most early fossil bats (~50 million years old) had wing designs suited for aerially hawking and not 4 Prey Detection and Discrimination in Animal-Eating Bats 97 like those of modern gleaners (Simmons and Geisler 1998 ; Safi et al. 2005 ). Therefore, we suppose that while something akin to gleaning may have characterized proto-bats and the very earliest of bats, this trait may have been subsequently lost, at least as a primary means of prey capture, as bats evolved more sophisticated laryngeal echolocation and longer, narrower wings, and then gleaning evolved inde- pendently again multiple times (Simmons and Geisler 1998 ) (Figure 4.1 ). However, while this idea is supported by fossil evidence (Simmons and Geisler 1998 ) and phylogenetic trait reconstructions (Safi et al. -

Integrated Fossil and Molecular Data Reconstruct Bat Echolocation
Integrated fossil and molecular data reconstruct bat echolocation Mark S. Springer†‡, Emma C. Teeling†§, Ole Madsen¶, Michael J. Stanhope§ʈ, and Wilfried W. de Jong¶** †Department of Biology, University of California, Riverside, CA 92521; §Queen’s University of Belfast, Biology and Biochemistry, 97 Lisburn Road, Belfast BT9 7BL, United Kingdom; ¶Department of Biochemistry, University of Nijmegen, P.O. Box 9101, 6500 HB Nijmegen, The Netherlands; ʈBioinformatics, SmithKline Beecham Pharmaceuticals, 1250 South Collegeville Road, UP1345, Collegeville, PA 19426; and **Institute for Biodiversity and Ecosystem Dynamics, 1090 GT Amsterdam, The Netherlands Edited by David Pilbeam, Harvard University, Cambridge, MA, and approved March 26, 2001 (received for review November 21, 2000) Molecular and morphological data have important roles in illumi- tulates a sister-group relationship between flying lemurs and bats nating evolutionary history. DNA data often yield well resolved (6). Although some authors have questioned bat monophyly phylogenies for living taxa, but are generally unattainable for based on evidence from the penis and nervous system (7, 8), the fossils. A distinct advantage of morphology is that some types of bulk of morphological data supports bat monophyly (9). Among morphological data may be collected for extinct and extant taxa. living Chiroptera, morphological data provide strong support for Fossils provide a unique window on evolutionary history and may the reciprocal monophyly of megachiropterans (Old World fruit preserve combinations -

Evolution of Body Mass in Bats: Insights from a Large Supermatrix Phylogeny
Journal of Mammalian Evolution https://doi.org/10.1007/s10914-018-9447-8 ORIGINAL PAPER Evolution of Body Mass in Bats: Insights from a Large Supermatrix Phylogeny Reyna Leticia Moyers Arévalo1 & Lucila I. Amador1 & Francisca C. Almeida2 & Norberto P. Giannini1,3,4 # Springer Science+Business Media, LLC, part of Springer Nature 2018 Abstract Bats are atypical small mammals. Size is crucial for bats because it affects most aerodynamic variables and several key echolocation parameters. In turn, scaling relationships of both flight and echolocation have been suggested to constrain bat body size evolution. Previous studies have found a large phylogenetic effect and the inclusion of early Eocene fossil bats contributed to recover idiosyncratic body size change patterns in bats. Here, we test these previous hypotheses of bat body size evolution using a large, comprehensive supermatrix phylogeny (+800 taxa) to optimize body size and examine changes reconstructed along branches. Our analysis provides evidence of rapid stem phyletic nanism, an ancestral value stabilized at 12 g for crown-clade Chiroptera followed by backbone stasis, low-magnitude changes inside established families, and massive body size increase at accelerated rate in pteropodid subclades. Total variation amount explained by pteropodid subclades was 86.3%, with most changes reconstructed as phyletic increases but also apomorphic decreases. We evaluate these macroevolutionary patterns in light of the constraints hypothesis, and in terms of both neutral and adaptive evolutionary models. The reconstructed macroevo- lution of bat body size led us to propose that echolocation and flight work as successive, nested constraints limiting bat evolution along the body size scale. Keywords Chiroptera . -

The Evolution and Development of Chiropteran Flight
University of New Hampshire University of New Hampshire Scholars' Repository Honors Theses and Capstones Student Scholarship Spring 2020 The Evolution and Development of Chiropteran Flight Emmaline Willis University of New Hampshire Follow this and additional works at: https://scholars.unh.edu/honors Part of the Developmental Biology Commons, Evolution Commons, Other Ecology and Evolutionary Biology Commons, and the Zoology Commons Recommended Citation Willis, Emmaline, "The Evolution and Development of Chiropteran Flight" (2020). Honors Theses and Capstones. 538. https://scholars.unh.edu/honors/538 This Senior Honors Thesis is brought to you for free and open access by the Student Scholarship at University of New Hampshire Scholars' Repository. It has been accepted for inclusion in Honors Theses and Capstones by an authorized administrator of University of New Hampshire Scholars' Repository. For more information, please contact [email protected]. Willis 1 Emmaline Willis Zool Thesis Advisor- Bolker 9/25/19 The Evolution and Development of Chiropteran Flight Intro Flying is thought of as a reserved skill used by very few animals throughout history. These animals may be the birds outside your window, the pterodactyl in a children’s book, or the insects buzzing about on a hot day. There are also thousands of bats flapping around in the dark all around the world. Exploring the sky, bats are the only mammal capable of true powered flight (Gunnel and Simmons 2005). Bats are an extraordinarily diverse group of mammals. Chiroptera, the order name for this group of mammals, has over one thousand species (Hill, 2018). In fact, the number may be closer to 1,400 species (K. -

Flying Primates Revisited: DNA Hybridization with Fractionated, GC-Enriched DNA
South African Journal of Science Vol. 91 September 1995 JOHN T. ROBINSON FESTSCHRIFT 477 :Flying primates revisited: DNA hybridization with fractionated, GC-enriched DNA John D. PettigreW' and John A.W. Kirsch2 'Vision, Touch and Hearing Research Centre, University of Queensland, Queensland 4072, Australia and 2Department of Zoology and Zoology Museum, University of Wisconsin, Madison 53705, USA. Molecular studies have been brought to bear on the primates from other mammalian orders. I I The finding of all of debate over the nature of the evolutionary links between these shared derived brain characters in flying foxes and pri primates and bats. The present investigation supports the mates, but not in microbats, resurrects the old notions about fly view that base composition is important in understanding ing primates. It is very hard to explain how these sophisticated phylogenetic relationships. Our discovery that some bats brain characters appeared independently in flying foxes and pri have extreme base compositional biases in their DNA com mates. The selective forces that would bring about such simila pared to others may help in understanding the origin of rity in functionally-separate brain systems are obscure. More these biases. over, with time, more and more examples of shared derived brain characters have been found linking primates and flying foxes A link between primates and bats has a long history. Linnaeus' (see review of this by Pettigrew(2). In other words, it is becoming placed bats into his Order Primates on the basis of anatomical increasingly unparsimonious to suggest that the primate brain features like the pendulous penis and axillary breasts that are characters that are also found in flying foxes have evolved inde shared by primates and some bats. -

Bats, That Is Yet to Be Seen
Janice Pease (315)328-5793 [email protected] 130 Beebe Rd, Potsdam, N.Y. 13676 August 23, 2018 Via Email Honorable Kathleen H. Burgess, Secretary to the PSC Re: Case 16- F-0268, Application of Atlantic Wind LLC for a certificate of Environmental Compatibility and Public Need Pursuant to Article 10 for Construction of the North Ridge Wind Energy Project in the Towns of Parishville and Hopkinton, St. Lawrence County. Dear Secretary Burgess: Industrial wind is devastating to the bat populations, adding to the many factors which play a role in reducing their numbers worldwide. While the wind industry likes to suggest that the advantage turbines provide to help reduce climate change (inadvertently benefitting all creatures) far outweighs their negative impact on the bats, that is yet to be seen. The data is simply not available to calculate the environmental/financial net losses accurately. With industrial wind’s intermittent and unreliable energy, the advantages are not nearly as the wind lobbyists suggest. The fragmentation/depletion of critical habitat due to wind turbines massive land use affects all animal species reliant on that space, ricocheting down the food chain. The loss of habitat as well as loss of carbon-sinks make the industrial turbine a very unlikely savior for any species. The net loss has simply not been calculated. Farmers are easily drawn into the debate by hosting these “farms”, while receiving large financial payouts. These same farmers are seemingly unaware of the immense benefit that bats provide by eating insect pests, saving farmers billions/year. The weakening of the local ecosystems will most certainly result in lower crop yields as well as contribute to financial losses as well. -

The Soft Explosive Model of Placental Mammal Evolution
bioRxiv preprint doi: https://doi.org/10.1101/251520; this version posted January 22, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The soft explosive model of placental mammal evolution Matthew J. Phillips*,1 and Carmelo Fruciano1 1School of Earth, Environmental and Biological Sciences, Queensland University of Technology, Brisbane, Australia *Corresponding author: E-mail: [email protected] Abstract Recent molecular dating estimates for placental mammals echo fossil inferences for an explosive interordinal diversification, but typically place this event some 10-20 million years earlier than the Paleocene fossils, among apparently more “primitive” mammal faunas. However, current models of molecular evolution do not adequately account for parallel rate changes, and result in dramatic divergence underestimates for large, long-lived mammals such as whales and hominids. Calibrating among these taxa shifts the rate model errors deeper in the tree, inflating interordinal divergence estimates. We employ simulations based on empirical rate variation, which show that this “error-shift inflation” can explain previous molecular dating overestimates relative to fossil inferences. Molecular dating accuracy is substantially improved in the simulations by focusing on calibrations for taxa that retain plesiomorphic life-history characteristics. Applying this strategy to the empirical data favours the soft explosive model of placental evolution, in line with traditional palaeontological interpretations – a few Cretaceous placental lineages give rise to a rapid interordinal diversification following the 66 Ma Cretaceous-Paleogene boundary mass extinction. -

1509831112.Full.Pdf
Chemical, experimental, and morphological evidence for diagenetically altered melanin in exceptionally preserved fossils Caitlin Collearya,b, Andrei Dolocanc, James Gardnerd, Suresh Singha, Michael Wuttkee, Renate Rabensteinf, Jörg Habersetzerf, Stephan Schaalf, Mulugeta Fesehag, Matthew Clemensh, Bonnie F. Jacobsh, Ellen D. Curranoi, Louis L. Jacobsh, Rene Lyng Sylvestersenj, Sarah E. Gabbottk, and Jakob Vinthera,d,l,1 aSchool of Earth Sciences, University of Bristol, Bristol BS8 1RJ, United Kingdom; bDepartment of Geosciences, Virginia Polytechnic Institute and State University, Blacksburg, VA 24060; cTexas Materials Institute, University of Texas at Austin, Austin, TX 78712; dJackson School of Geosciences, University of Texas at Austin, Austin, TX 78712; eDepartment for the Conservation of the Cultural Heritage of Rhineland-Palatinate, 55116 Mainz, Germany; fDepartment of Palaeoanthropology and Messel Research, Senckenberg Research Institute, 60325 Frankfurt am Main, Germany; gPaleoanthropology and Paleoenvironment Program, School of Earth Sciences, College of Natural Sciences, Addis Ababa University, Addis Ababa, Ethiopia; hRoy M. Huffington Department of Earth Sciences, Southern Methodist University, Dallas, TX 75275; iDepartments of Botany and Geology & Geophysics, University of Wyoming, Laramie, WY 82071; jDivision of Natural History, MuseÒum, 7800 Skive, Denmark; kDepartment of Geology, University of Leicester, Leicester LE1 7RH, United Kingdom; and lSchool of Biological Sciences, University of Bristol, Bristol BS8 1TQ, United Kingdom Edited by Donald E. Canfield, Institute of Biology and Nordic Center for Earth Evolution, University of Southern Denmark, Odense M., Denmark, and approved August 26, 2015 (received for review May 19, 2015) In living organisms, color patterns, behavior, and ecology are closely evidence distinguishes pure eumelanin and phaeomelanin in linked. Thus, detection of fossil pigments may permit inferences turtles (14), and phaeomelanin has been tentatively reported in about important aspects of ancient animal ecology and evolution.