The Soft Explosive Model of Placental Mammal Evolution
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The World at the Time of Messel: Conference Volume
T. Lehmann & S.F.K. Schaal (eds) The World at the Time of Messel - Conference Volume Time at the The World The World at the Time of Messel: Puzzles in Palaeobiology, Palaeoenvironment and the History of Early Primates 22nd International Senckenberg Conference 2011 Frankfurt am Main, 15th - 19th November 2011 ISBN 978-3-929907-86-5 Conference Volume SENCKENBERG Gesellschaft für Naturforschung THOMAS LEHMANN & STEPHAN F.K. SCHAAL (eds) The World at the Time of Messel: Puzzles in Palaeobiology, Palaeoenvironment, and the History of Early Primates 22nd International Senckenberg Conference Frankfurt am Main, 15th – 19th November 2011 Conference Volume Senckenberg Gesellschaft für Naturforschung IMPRINT The World at the Time of Messel: Puzzles in Palaeobiology, Palaeoenvironment, and the History of Early Primates 22nd International Senckenberg Conference 15th – 19th November 2011, Frankfurt am Main, Germany Conference Volume Publisher PROF. DR. DR. H.C. VOLKER MOSBRUGGER Senckenberg Gesellschaft für Naturforschung Senckenberganlage 25, 60325 Frankfurt am Main, Germany Editors DR. THOMAS LEHMANN & DR. STEPHAN F.K. SCHAAL Senckenberg Research Institute and Natural History Museum Frankfurt Senckenberganlage 25, 60325 Frankfurt am Main, Germany [email protected]; [email protected] Language editors JOSEPH E.B. HOGAN & DR. KRISTER T. SMITH Layout JULIANE EBERHARDT & ANIKA VOGEL Cover Illustration EVELINE JUNQUEIRA Print Rhein-Main-Geschäftsdrucke, Hofheim-Wallau, Germany Citation LEHMANN, T. & SCHAAL, S.F.K. (eds) (2011). The World at the Time of Messel: Puzzles in Palaeobiology, Palaeoenvironment, and the History of Early Primates. 22nd International Senckenberg Conference. 15th – 19th November 2011, Frankfurt am Main. Conference Volume. Senckenberg Gesellschaft für Naturforschung, Frankfurt am Main. pp. 203. -

Special Publications Museum of Texas Tech University Number 63 18 September 2014
Special Publications Museum of Texas Tech University Number 63 18 September 2014 List of Recent Land Mammals of Mexico, 2014 José Ramírez-Pulido, Noé González-Ruiz, Alfred L. Gardner, and Joaquín Arroyo-Cabrales.0 Front cover: Image of the cover of Nova Plantarvm, Animalivm et Mineralivm Mexicanorvm Historia, by Francisci Hernández et al. (1651), which included the first list of the mammals found in Mexico. Cover image courtesy of the John Carter Brown Library at Brown University. SPECIAL PUBLICATIONS Museum of Texas Tech University Number 63 List of Recent Land Mammals of Mexico, 2014 JOSÉ RAMÍREZ-PULIDO, NOÉ GONZÁLEZ-RUIZ, ALFRED L. GARDNER, AND JOAQUÍN ARROYO-CABRALES Layout and Design: Lisa Bradley Cover Design: Image courtesy of the John Carter Brown Library at Brown University Production Editor: Lisa Bradley Copyright 2014, Museum of Texas Tech University This publication is available free of charge in PDF format from the website of the Natural Sciences Research Laboratory, Museum of Texas Tech University (nsrl.ttu.edu). The authors and the Museum of Texas Tech University hereby grant permission to interested parties to download or print this publication for personal or educational (not for profit) use. Re-publication of any part of this paper in other works is not permitted without prior written permission of the Museum of Texas Tech University. This book was set in Times New Roman and printed on acid-free paper that meets the guidelines for per- manence and durability of the Committee on Production Guidelines for Book Longevity of the Council on Library Resources. Printed: 18 September 2014 Library of Congress Cataloging-in-Publication Data Special Publications of the Museum of Texas Tech University, Number 63 Series Editor: Robert J. -

Neoichnology of Bats: Morphological, Ecological, and Phylogenetic Influences on Terrestrial Behavior and Trackmaking Ability Within the Chiroptera
NEOICHNOLOGY OF BATS: MORPHOLOGICAL, ECOLOGICAL, AND PHYLOGENETIC INFLUENCES ON TERRESTRIAL BEHAVIOR AND TRACKMAKING ABILITY WITHIN THE CHIROPTERA BY MATTHEW FRAZER JONES Submitted to the graduate degree program in Geology and the Graduate Faculty of the University of Kansas in partial fulfillment of the requirements for the degree of Master of Science. Advisory Committee: ______________________________ Chairperson Stephen T. Hasiotis ______________________________ Co-chair David A. Burnham ______________________________ Robert M. Timm Date Defended: April 8, 2016 The Thesis Committee for MATTHEW FRAZER JONES certifies that this is the approved version of the following thesis: NEOICHNOLOGY OF BATS: MORPHOLOGICAL, ECOLOGICAL, AND PHYLOGENETIC INFLUENCES ON TERRESTRIAL BEHAVIOR AND TRACKMAKING ABILITY WITHIN THE CHIROPTERA ______________________________ Chairperson: Stephen T. Hasiotis ______________________________ Co-chairperson: David A. Burnham Date Approved: April 8, 2016 ii ABSTRACT Among living mammals, bats (Chiroptera) are second only to rodents in total number of species with over 1100 currently known. Extant bat species occupy many trophic niches and feeding habits, including frugivores (fruit eaters), insectivores (insect eaters), nectarivores (nectar and pollen-eaters), carnivores (predators of small terrestrial vertebrates), piscivores (fish eaters), sanguinivores (blood eaters), and omnivores (eat animals and plant material). Modern bats also demonstrate a wide range of terrestrial abilities while feeding, including: (1) those that primarily feed at or near ground level, such as the common vampire bat (Desmodus rotundus) and the New Zealand short-tailed bat (Mystacina tuberculata); (2) those rarely observed to feed from or otherwise spend time on the ground; and (3) many intermediate forms that demonstrate terrestrial competency without an obvious ecological basis. The variation in chiropteran terrestrial ability has been hypothesized to be constrained by the morphology of the pelvis and hindlimbs into what are termed types 1, 2, and 3 bats. -
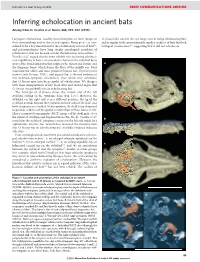
Inferring Echolocation in Ancient Bats Arising From: N
NATURE | Vol 466 | 19 August 2010 BRIEF COMMUNICATIONS ARISING Inferring echolocation in ancient bats Arising from: N. Veselka et al. Nature 463, 939–942 (2010) Laryngeal echolocation, used by most living bats to form images of O. finneyi falls outside the size range seen in living echolocating bats their surroundings and to detect and capture flying prey1,2, is con- and is similar to the proportionally smaller cochleae of bats that lack sidered to be a key innovation for the evolutionary success of bats2,3, laryngeal echolocation4,8, suggesting that it did not echolocate. and palaeontologists have long sought osteological correlates of echolocation that can be used to infer the behaviour of fossil bats4–7. Veselka et al.8 argued that the most reliable trait indicating echoloca- tion capabilities in bats is an articulation between the stylohyal bone (part of the hyoid apparatus that supports the throat and larynx) and a the tympanic bone, which forms the floor of the middle ear. They examined the oldest and most primitive known bat, Onychonycteris finneyi (early Eocene, USA4), and argued that it showed evidence of this stylohyal–tympanic articulation, from which they concluded that O. finneyi may have been capable of echolocation. We disagree with their interpretation of key fossil data and instead argue that O. finneyi was probably not an echolocating bat. The holotype of O. finneyi shows the cranial end of the left stylohyal resting on the tympanic bone (Fig. 1c–e). However, the stylohyal on the right side is in a different position, the tip of the stylohyal extends beyond the tympanic on both sides of the skull, and both tympanics are crushed. -

BIO 313 ANIMAL ECOLOGY Corrected
NATIONAL OPEN UNIVERSITY OF NIGERIA SCHOOL OF SCIENCE AND TECHNOLOGY COURSE CODE: BIO 314 COURSE TITLE: ANIMAL ECOLOGY 1 BIO 314: ANIMAL ECOLOGY Team Writers: Dr O.A. Olajuyigbe Department of Biology Adeyemi Colledge of Education, P.M.B. 520, Ondo, Ondo State Nigeria. Miss F.C. Olakolu Nigerian Institute for Oceanography and Marine Research, No 3 Wilmot Point Road, Bar-beach Bus-stop, Victoria Island, Lagos, Nigeria. Mrs H.O. Omogoriola Nigerian Institute for Oceanography and Marine Research, No 3 Wilmot Point Road, Bar-beach Bus-stop, Victoria Island, Lagos, Nigeria. EDITOR: Mrs Ajetomobi School of Agricultural Sciences Lagos State Polytechnic Ikorodu, Lagos 2 BIO 313 COURSE GUIDE Introduction Animal Ecology (313) is a first semester course. It is a two credit unit elective course which all students offering Bachelor of Science (BSc) in Biology can take. Animal ecology is an important area of study for scientists. It is the study of animals and how they related to each other as well as their environment. It can also be defined as the scientific study of interactions that determine the distribution and abundance of organisms. Since this is a course in animal ecology, we will focus on animals, which we will define fairly generally as organisms that can move around during some stages of their life and that must feed on other organisms or their products. There are various forms of animal ecology. This includes: • Behavioral ecology, the study of the behavior of the animals with relation to their environment and others • Population ecology, the study of the effects on the population of these animals • Marine ecology is the scientific study of marine-life habitat, populations, and interactions among organisms and the surrounding environment including their abiotic (non-living physical and chemical factors that affect the ability of organisms to survive and reproduce) and biotic factors (living things or the materials that directly or indirectly affect an organism in its environment). -

Supplemental File 1: Addressing Claims of “Zombie” Lineages on Phillips’ (2016) Timetree
Supplemental File 1: Addressing claims of “zombie” lineages on Phillips’ (2016) timetree The soft explosive model of placental mammal evolution Matthew J. Phillips*,1 and Carmelo Fruciano1 1School of Earth, Environmental and Biological Sciences, Queensland University of Technology, Brisbane, Australia *Corresponding author: E-mail: [email protected] Contents Addressing claims of “zombie” lineages on Phillips’ (2016) timetree ................................................... 1 Incorrect or poorly supported fossil placements ................................................................................. 1 Figure S1 ............................................................................................................................................ 4 Table S1 .............................................................................................................................................. 6 References .............................................................................................................................................. 7 Addressing claims of “zombie lineages” on Phillips’ (2016) timetree Phillips [1] found extreme divergence underestimation among large, long-lived taxa that were not calibrated, and argued that calibrating these taxa instead shifted the impact of the underlying rate model misspecification to inflating dates deeper in the tree. To avoid this “error-shift inflation”, Phillips [1] first inferred divergences with dos Reis et al.’s [2] calibrations, most of which are set among taxa -

Hyaenodontidae (Creodonta, Mammalia) and the Position of Systematics in Evolutionary Biology
Hyaenodontidae (Creodonta, Mammalia) and the Position of Systematics in Evolutionary Biology by Paul David Polly B.A. (University of Texas at Austin) 1987 A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Paleontology in the GRADUATE DIVISION of the UNIVERSITY of CALIFORNIA at BERKELEY Committee in charge: Professor William A. Clemens, Chair Professor Kevin Padian Professor James L. Patton Professor F. Clark Howell 1993 Hyaenodontidae (Creodonta, Mammalia) and the Position of Systematics in Evolutionary Biology © 1993 by Paul David Polly To P. Reid Hamilton, in memory. iii TABLE OF CONTENTS Introduction ix Acknowledgments xi Chapter One--Revolution and Evolution in Taxonomy: Mammalian Classification Before and After Darwin 1 Introduction 2 The Beginning of Modern Taxonomy: Linnaeus and his Predecessors 5 Cuvier's Classification 10 Owen's Classification 18 Post-Darwinian Taxonomy: Revolution and Evolution in Classification 24 Kovalevskii's Classification 25 Huxley's Classification 28 Cope's Classification 33 Early 20th Century Taxonomy 42 Simpson and the Evolutionary Synthesis 46 A Box Model of Classification 48 The Content of Simpson's 1945 Classification 50 Conclusion 52 Acknowledgments 56 Bibliography 56 Figures 69 Chapter Two: Hyaenodontidae (Creodonta, Mammalia) from the Early Eocene Four Mile Fauna and Their Biostratigraphic Implications 78 Abstract 79 Introduction 79 Materials and Methods 80 iv Systematic Paleontology 80 The Four Mile Fauna and Wasatchian Biostratigraphic Zonation 84 Conclusion 86 Acknowledgments 86 Bibliography 86 Figures 87 Chapter Three: A New Genus Eurotherium (Creodonta, Mammalia) in Reference to Taxonomic Problems with Some Eocene Hyaenodontids from Eurasia (With B. Lange-Badré) 89 Résumé 90 Abstract 90 Version française abrégéé 90 Introduction 93 Acknowledgments 96 Bibliography 96 Table 3.1: Original and Current Usages of Genera and Species 99 Table 3.2: Species Currently Included in Genera Discussed in Text 101 Chapter Four: The skeleton of Gazinocyon vulpeculus n. -

Molecular Phylogeny of Mobatviruses (Hantaviridae) in Myanmar and Vietnam
viruses Article Molecular Phylogeny of Mobatviruses (Hantaviridae) in Myanmar and Vietnam Satoru Arai 1, Fuka Kikuchi 1,2, Saw Bawm 3 , Nguyễn Trường Sơn 4,5, Kyaw San Lin 6, Vương Tân Tú 4,5, Keita Aoki 1,7, Kimiyuki Tsuchiya 8, Keiko Tanaka-Taya 1, Shigeru Morikawa 9, Kazunori Oishi 1 and Richard Yanagihara 10,* 1 Infectious Disease Surveillance Center, National Institute of Infectious Diseases, Tokyo 162-8640, Japan; [email protected] (S.A.); [email protected] (F.K.); [email protected] (K.A.); [email protected] (K.T.-T.); [email protected] (K.O.) 2 Department of Chemistry, Faculty of Science, Tokyo University of Science, Tokyo 162-8601, Japan 3 Department of Pharmacology and Parasitology, University of Veterinary Science, Yezin, Nay Pyi Taw 15013, Myanmar; [email protected] 4 Institute of Ecology and Biological Resources, Vietnam Academy of Science and Technology, Hanoi, Vietnam; [email protected] (N.T.S.); [email protected] (V.T.T.) 5 Graduate University of Science and Technology, Vietnam Academy of Science and Technology, Hanoi, Vietnam 6 Department of Aquaculture and Aquatic Disease, University of Veterinary Science, Yezin, Nay Pyi Taw 15013, Myanmar; [email protected] 7 Department of Liberal Arts, Faculty of Science, Tokyo University of Science, Tokyo 162-8601, Japan 8 Laboratory of Bioresources, Applied Biology Co., Ltd., Tokyo 107-0062, Japan; [email protected] 9 Department of Veterinary Science, National Institute of Infectious Diseases, Tokyo 162-8640, Japan; [email protected] 10 Pacific Center for Emerging Infectious Diseases Research, John A. -

103 the EVOLUTION of BATSI Leigh Van Valen Department Of
103 THE EVOLUTIONOF BATSI Leigh Van Valen Department of Biology University of Chicago 1103 East 57th Street Chicago, Illinois 60637 Received August 30, 1978; June 8, L979 Abstract: I devel-op the first explicitly Justified phylogeny of the known farnil-les of bats, usj.ng all available characters. This phylogeny permits the adaptive evolution of the order to be outlined. Two main clades emerge within the Microchiroptera and are cal-led new infraorders, Vespertilionia and Phyllostomatia. In each infraorder there is a series of grades of progressively stronger fi-ight, and there is a radiation of diet within the Phyllostomatia. Parallel evolution is extensive. Most grades stlll exist, presumably by a poorly understood partitioning of the resource space. The Megachiroptera nay have originated in the l-ate Ollgocene or early Miocene from surviving mernbers of the Eochiroptera (new suborder). The Kerivoul,ldae, Myzopodidae, Thyropteridae, and Furlpterldae are placed in the Natalidae, and the lcaronycterididae in the PaLaeochiropterygidae. The features of an ancestral bat are predicted. Bat origins are poorly known but may be found in Paleocene members of the Adapisoricidae. ,r** Bats constitute the second largest order of marnmalsand have a readily decipherable adaptive history, at least compared with the Rodentia and Insectivora. Nevertheless, there is no treatment of this adaptive hlstory nor even a general phyl-ogeny, which must form its foundation. I noticed this l-ack when revising a course on the paleobiology of mauunalsand undertook to remedy it. Although I sttll lack much familiarity with bats, the resul-t may be of more general interest. ORIGIN OF BATS One may hypothesize ttrat bats dld originate, but it is harder to go beyond this. -

Jurnal Riset Veteriner Indonesia Journal of the Indonesian Veterinary Research P-ISSN: 2614-0187, E-ISSN:2615-2835 Volume 3 No
Jurnal Riset Veteriner Indonesia Journal of the Indonesian Veterinary Research P-ISSN: 2614-0187, E-ISSN:2615-2835 Volume 3 No. 1 (Januari 2019), pp. 1-9 journal.unhas.ac.id/index.php/jrvi/ This woks is licenced under a Creative Commons Attribution 4.0 International License. R eview Article Bats Oxidative Stress Defense (Pertahanan Stres Oksidatif pada Kelelawar) 1 1 1 Desrayni Hanadhita , Aryani Sismin Satjaningtyas , Srihadi Agungpriyono * 1Department of Anatomy Physiology and Pharmacology, Faculty of Veterinary Medicine, IPB University Jl. Agatis, Kampus IPB Dramaga, Bogor 16680, Indonesia *Corresponding authors: Srihadi Agungpriyono ([email protected]) Abstract Antioxidants and free radicals have long been known to be the main factors in the occurrence of degenerative diseases. Various studies related to antioxidants and free radicals which have implications for oxidative stress have increased in the last decade. Knowledge of stress oxidative physiology in various animals help in understanding the pathophysiology of diseases associated with oxidative stress. Bats are claimed to be the best known animals in term of survival compared to other mammals. Bats are reported to produce low reactive oxygen species (ROS) but high endogenous antioxidants that can prevent oxidative stress. Bats high defense against oxidative stress has implications for their extreme longevity, the role as a reservoir of viruses, and the potential as experimental animals. Key words: aging, animal model, antioxidant, comparative physiology, free radical Copyright © 2019 JRVI. All rights reserved. Introduction Oxidative stress has been a scourge for the health world for more than three decades (Sies, 2015). Research on oxidative stress status related to disease pathogenesis is increasing every year. -

The Phylogenetic Roots of Human Lethal Violence José María Gómez1,2, Miguel Verdú3, Adela González-Megías4 & Marcos Méndez5
LETTER doi:10.1038/nature19758 The phylogenetic roots of human lethal violence José María Gómez1,2, Miguel Verdú3, Adela González-Megías4 & Marcos Méndez5 The psychological, sociological and evolutionary roots of 600 human populations, ranging from the Palaeolithic era to the present conspecific violence in humans are still debated, despite attracting (Supplementary Information section 9c). The level of lethal violence the attention of intellectuals for over two millennia1–11. Here we was defined as the probability of dying from intraspecific violence propose a conceptual approach towards understanding these roots compared to all other causes. More specifically, we calculated the level based on the assumption that aggression in mammals, including of lethal violence as the percentage, with respect to all documented humans, has a significant phylogenetic component. By compiling sources of mortality, of total deaths due to conspecifics (these sources of mortality from a comprehensive sample of mammals, were infanticide, cannibalism, inter-group aggression and any other we assessed the percentage of deaths due to conspecifics and, type of intraspecific killings in non-human mammals; war, homicide, using phylogenetic comparative tools, predicted this value for infanticide, execution, and any other kind of intentional conspecific humans. The proportion of human deaths phylogenetically killing in humans). predicted to be caused by interpersonal violence stood at 2%. Lethal violence is reported for almost 40% of the studied mammal This value was similar to the one phylogenetically inferred for species (Supplementary Information section 9a). This is probably the evolutionary ancestor of primates and apes, indicating that a an underestimation, because information is not available for many certain level of lethal violence arises owing to our position within species. -

Genomic Analysis Reveals Hidden Biodiversity Within Colugos, the Sister Group to Primates Victor C
Washington University School of Medicine Digital Commons@Becker Open Access Publications 2016 Genomic analysis reveals hidden biodiversity within colugos, the sister group to primates Victor C. Mason Texas A & M University - College Station Gang Li Texas A & M University - College Station Patrick Minx Washington University School of Medicine in St. Louis Jürgen Schmitz University of Münster Gennady Churakov University of Münster See next page for additional authors Follow this and additional works at: https://digitalcommons.wustl.edu/open_access_pubs Recommended Citation Mason, Victor C.; Li, Gang; Minx, Patrick; Schmitz, Jürgen; Churakov, Gennady; Doronina, Liliya; Melin, Amanda D.; Dominy, Nathaniel J.; Lim, Norman T-L; Springer, Mark S.; Wilson, Richard K.; Warren, Wesley C.; Helgen, Kristofer M.; and Murphy, William J., ,"Genomic analysis reveals hidden biodiversity within colugos, the sister group to primates." Science Advances.2,8. e1600633. (2016). https://digitalcommons.wustl.edu/open_access_pubs/5209 This Open Access Publication is brought to you for free and open access by Digital Commons@Becker. It has been accepted for inclusion in Open Access Publications by an authorized administrator of Digital Commons@Becker. For more information, please contact [email protected]. Authors Victor C. Mason, Gang Li, Patrick Minx, Jürgen Schmitz, Gennady Churakov, Liliya Doronina, Amanda D. Melin, Nathaniel J. Dominy, Norman T-L Lim, Mark S. Springer, Richard K. Wilson, Wesley C. Warren, Kristofer M. Helgen, and William J. Murphy This open access publication is available at Digital Commons@Becker: https://digitalcommons.wustl.edu/open_access_pubs/5209 RESEARCH ARTICLE ZOOLOGICAL POPULATION GENETICS 2016 © The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science.