Generated by SRI International Pathway Tools Version 25.0, Authors S
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Review Article RNA Degradation in Staphylococcus Aureus: Diversity of Ribonucleases and Their Impact
Hindawi Publishing Corporation International Journal of Genomics Volume 2015, Article ID 395753, 12 pages http://dx.doi.org/10.1155/2015/395753 Review Article RNA Degradation in Staphylococcus aureus: Diversity of Ribonucleases and Their Impact Rémy A. Bonnin and Philippe Bouloc Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, UniversiteParis-Sud,Universit´ eParis-Saclay,91400Orsay,France´ Correspondence should be addressed to Remy´ A. Bonnin; [email protected] Received 6 January 2015; Accepted 4 March 2015 Academic Editor: Martine A. Collart Copyright © 2015 R. A. Bonnin and P. Bouloc. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The regulation of RNA decay is now widely recognized as having a central role in bacterial adaption to environmental stress.Here we present an overview on the diversity of ribonucleases (RNases) and their impact at the posttranscriptional level in the human pathogen Staphylococcus aureus. RNases in prokaryotes have been mainly studied in the two model organisms Escherichia coli and Bacillus subtilis. Based on identified RNases in these two models, putative orthologs have been identified in S. aureus.Themain staphylococcal RNases involved in the processing and degradation of the bulk RNA are (i) endonucleases RNase III and RNase Y and (ii) exonucleases RNase J1/J2 and PNPase, having 5 to 3 and 3 to 5 activities, respectively. The diversity and potential roles of each RNase and of Hfq and RppH are discussed in the context of recent studies, some of which are based on next-generation sequencing technology. -

Generated by SRI International Pathway Tools Version 25.0, Authors S
An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000238675-HmpCyc: Bacillus smithii 7_3_47FAA Cellular Overview Connections between pathways are omitted for legibility. -
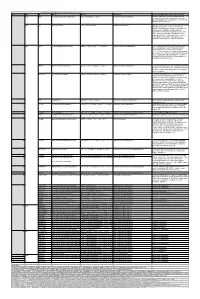
When the Reaction Is
Table S3. iJL1678-ME model modification (blocked reactions) Iter. Cat. ID Name Formula Subsystem Comments (When the reaction is turned on) 1 bp2 EDD 6-phosphogluconate dehydratase 6pgc_c⇌2ddg6p_c + h2o_c Pentose Phosphate Pathway Create a major effect of steep acetate overflow elevation in high growth. Comparing to the main glycolytic pathway, it is metabolicly less efficient but proteomicly more efficient. bp1 ICL Isocitrate lyase icit_c→glx_c + succ_c Anaplerotic Reactions Bypass for the main TCA cycle pathways from turning isocitrate to succinate, when ICL is turned on, Isocitrate dehydrogenase(ICDHyr), 2-Oxogluterate dehydrogenase(AKGDH) and Succinyl-CoA synthetase (ATP-forming,SUCOAS) would reduce. Ref. (1) and (2) shows that this reaction is off in higher growth. Ref. (3) shows that this reaction is converging to being off when the dynamic of respiration using enzyme kinetics is simulated. 2 bp1 ABTA 4-aminobutyrate transaminase 4abut_c + akg_c⇌glu__L_c + sucsal_c Arginine and Proline Metabolism Another backup pathway of succinate production, from 2-Oxoglutarate (akg). Respiration would be induced when it is on, since the flux through ETC(CYTBO3_4pp and ATPS4rpp) would increase. As it requires the co-factor pyridoxal 5'-phosphate(2−) to get catalyzed(4), indicating that this reaction is regulated by the flux of other reactions(pyridoxal 5'- phosphate(2-) production, etc.). 3 GLYAT Glycine C-acetyltransferase accoa_c + gly_c⇌2aobut_c + coa_c Glycine and Serine Metabolism A reaction that back up for the respiration. Reactions fluxes in TCA cycle would drop when this reaction is turned on. It also requires pyridoxal 5'-phosphate(2−) for the regulation. 4 NADTRHD NAD transhydrogenase nad_c + nadph_c⇌nadh_c + nadp_c Oxidative Phosphorylation A reaction that would make the transition between NAD and NADP metabolically more efficient. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Tese Jucimar Zacaria.Pdf (4.725Mb)
UNIVERSIDADE DE CAXIAS DO SUL CENTRO DE CIÊNCIAS BIOLÓGICAS E DA SAÚDE INSTITUTO DE BIOTECNOLOGIA PROGRAMA DE PÓS-GRADUAÇÃO EM BIOTECNOLOGIA DIVERSIDADE, CLONAGEM E CARACTERIZAÇÃO DE NUCLEASES EXTRACELULARES DE Aeromonas spp. JUCIMAR ZACARIA CAXIAS DO SUL 2016 JUCIMAR ZACARIA DIVERSIDADE, CLONAGEM E CARACTERIZAÇÃO DE NUCLEASES EXTRACELULARES DE Aeromonas spp. Tese apresentada ao programa de Pós- graduação em Biotecnologia da Universidade de Caxias do Sul, visando à obtenção de grau de Doutor em Biotecnologia. Orientador: Dr. Sergio Echeverrigaray Co-orientador: Dra. Ana Paula Longaray Delamare Caxias do Sul 2016 ii Z13d Zacaria, Jucimar Diversidade, clonagem e caracterização de nucleases extracelulares de Aeromonas spp. / Jucimar Zacaria. – 2016. 258 f.: il. Tese (Doutorado) - Universidade de Caxias do Sul, Programa de Pós- Graduação em Biotecnologia, 2016. Orientação: Sergio Echeverrigaray. Coorientação: Ana Paula Longaray Delamare. 1. Aeromas. 2. DNases extracelulares. 3. Termoestabilidade. 4. Dns. 5. Aha3441. I. Echeverrigaray, Sergio, orient. II. Delamare, Ana Paula Longaray, coorient. III. Título. Elaborado pelo Sistema de Geração Automática da UCS com os dados fornecidos pelo(a) autor(a). JUCIMAR ZACARIA DIVERSIDADE, CLONAGEM E CARACTERIZAÇÃO DE NUCLEASES EXTRACELULARES DE Aeromonas spp. Tese apresentada ao Programa de Pós-graduação em Biotecnologia da Universidade de Caxias do Sul, visando à obtenção do título de Doutor em Biotecnologia. Orientador: Prof. Dr. Sergio Echeverrigaray Laguna Co-orientadora: Profa. Dra. Ana Paula Longaray -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -

Generated by SRI International Pathway Tools Version 25.0, Authors S
Authors: Pallavi Subhraveti Ron Caspi Peter Midford Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000157595Cyc: Enterococcus faecium 1,231,410 Cellular Overview Connections between pathways are omitted for legibility. -
Generated by SRI International Pathway Tools Version 25.0, Authors S
Authors: Pallavi Subhraveti Ron Caspi Peter Midford Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000166655Cyc: Borreliella burgdorferi JD1 Cellular Overview Connections between pathways are omitted for legibility. -

Core Proteins in All 6 Lactobacillus Species Analyzed 1-Acyl-Sn-Glycerol
Core proteins in all 6 lactobacillus species analyzed 1-acyl-sn-glycerol-3-phosphate acyltransferase 16S rRNA m(5)C 967 methyltransferase 16S rRNA methyltransferase GidB 16S rRNA processing protein 23S rRNA methyltransferase 23S rRNA methyltransferase 3-hydroxy-3-methylglutaryl-coenzyme A reductase/3-hydroxy-3-methylglutaryl-coenzyme Auctase 30S ribosomal protein S1 30S ribosomal protein S10 30S ribosomal protein S11 30S ribosomal protein S12 30S ribosomal protein S13 30S ribosomal protein S16 30S ribosomal protein S17 30S ribosomal protein S18 30S ribosomal protein S19 30S ribosomal protein S2 30S ribosomal protein S20 30S ribosomal protein S21 30S ribosomal protein S3 30S ribosomal protein S4 30S ribosomal protein S5 30S ribosomal protein S7 30S ribosomal protein S8 30S ribosomal protein S9 4-oxalocrotonate tautomerase 4'-phosphopantetheinyl transferase 5-formyltetrahydrofolate cyclo-ligase 5'-methylthioadenosine nucleosidase / S-adenosylhomocysteine nucleosidase 50S ribosomal protein L1 50S ribosomal protein L10 50S ribosomal protein L11 50S ribosomal protein L12P 50S ribosomal protein L13 50S ribosomal protein L14P 50S ribosomal protein L15P 50S ribosomal protein L16 50S ribosomal protein L17P 50S ribosomal protein L18 50S ribosomal protein L19 50S ribosomal protein L2 50S ribosomal protein L20 50S ribosomal protein L21 50S ribosomal protein L22 50S ribosomal protein L23P 50S ribosomal protein L24P 50S ribosomal protein L27,ABC superfamily ATP binding cassette transporter, substrate binding protein 50S ribosomal protein L28 50S ribosomal -

All Enzymes in BRENDA™ the Comprehensive Enzyme Information System
All enzymes in BRENDA™ The Comprehensive Enzyme Information System http://www.brenda-enzymes.org/index.php4?page=information/all_enzymes.php4 1.1.1.1 alcohol dehydrogenase 1.1.1.B1 D-arabitol-phosphate dehydrogenase 1.1.1.2 alcohol dehydrogenase (NADP+) 1.1.1.B3 (S)-specific secondary alcohol dehydrogenase 1.1.1.3 homoserine dehydrogenase 1.1.1.B4 (R)-specific secondary alcohol dehydrogenase 1.1.1.4 (R,R)-butanediol dehydrogenase 1.1.1.5 acetoin dehydrogenase 1.1.1.B5 NADP-retinol dehydrogenase 1.1.1.6 glycerol dehydrogenase 1.1.1.7 propanediol-phosphate dehydrogenase 1.1.1.8 glycerol-3-phosphate dehydrogenase (NAD+) 1.1.1.9 D-xylulose reductase 1.1.1.10 L-xylulose reductase 1.1.1.11 D-arabinitol 4-dehydrogenase 1.1.1.12 L-arabinitol 4-dehydrogenase 1.1.1.13 L-arabinitol 2-dehydrogenase 1.1.1.14 L-iditol 2-dehydrogenase 1.1.1.15 D-iditol 2-dehydrogenase 1.1.1.16 galactitol 2-dehydrogenase 1.1.1.17 mannitol-1-phosphate 5-dehydrogenase 1.1.1.18 inositol 2-dehydrogenase 1.1.1.19 glucuronate reductase 1.1.1.20 glucuronolactone reductase 1.1.1.21 aldehyde reductase 1.1.1.22 UDP-glucose 6-dehydrogenase 1.1.1.23 histidinol dehydrogenase 1.1.1.24 quinate dehydrogenase 1.1.1.25 shikimate dehydrogenase 1.1.1.26 glyoxylate reductase 1.1.1.27 L-lactate dehydrogenase 1.1.1.28 D-lactate dehydrogenase 1.1.1.29 glycerate dehydrogenase 1.1.1.30 3-hydroxybutyrate dehydrogenase 1.1.1.31 3-hydroxyisobutyrate dehydrogenase 1.1.1.32 mevaldate reductase 1.1.1.33 mevaldate reductase (NADPH) 1.1.1.34 hydroxymethylglutaryl-CoA reductase (NADPH) 1.1.1.35 3-hydroxyacyl-CoA -

Structures of B. Subtilis Maturation Rnases Captured on 50S
Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs Stephanie Oerum, Tom Dendooven, Marjorie Catala, Laetitia Gilet, Clément Dégut, Aude Trinquier, Maxime Bourguet, Pierre Barraud, Sarah Cianferani, Ben Luisi, et al. To cite this version: Stephanie Oerum, Tom Dendooven, Marjorie Catala, Laetitia Gilet, Clément Dégut, et al.. Struc- tures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs. Molecular Cell, Elsevier, 2020, 80 (2), pp.227-236.e5. 10.1016/j.molcel.2020.09.008. hal-03015124 HAL Id: hal-03015124 https://hal.archives-ouvertes.fr/hal-03015124 Submitted on 19 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Structures of B. subtilis maturation RNases captured on 50S ribosome with pre-rRNAs Stephanie Oerum1, Tom Dendooven2, Marjorie Catala1, Laetitia Gilet1, Clément Dégut1, Aude Trinquier1, Maxime Bourguet3, Pierre Barraud1, Sarah Cianferani3, Ben F. Luisi2, Ciarán Condon1*, Carine Tisné1,4* 1Expression Génétique Microbienne, UMR 8261, CNRS, Université de Paris, Institut de Biologie Physico-Chimique (IBPC), 75005 Paris, France. 2Department of Biochemistry, University of Cambridge, Tennis Court Road, Cambridge CB2 1GA, U.K. 3Laboratoire de Spectrométrie de Masse BioOrganique, Université de Strasbourg, CNRS, IPHC UMR 7178, 67000 Strasbourg, France. -

Springer Handbook of Enzymes
Dietmar Schomburg Ida Schomburg (Eds.) Springer Handbook of Enzymes Alphabetical Name Index 1 23 © Springer-Verlag Berlin Heidelberg New York 2010 This work is subject to copyright. All rights reserved, whether in whole or part of the material con- cerned, specifically the right of translation, printing and reprinting, reproduction and storage in data- bases. The publisher cannot assume any legal responsibility for given data. Commercial distribution is only permitted with the publishers written consent. Springer Handbook of Enzymes, Vols. 1–39 + Supplements 1–7, Name Index 2.4.1.60 abequosyltransferase, Vol. 31, p. 468 2.7.1.157 N-acetylgalactosamine kinase, Vol. S2, p. 268 4.2.3.18 abietadiene synthase, Vol. S7,p.276 3.1.6.12 N-acetylgalactosamine-4-sulfatase, Vol. 11, p. 300 1.14.13.93 (+)-abscisic acid 8’-hydroxylase, Vol. S1, p. 602 3.1.6.4 N-acetylgalactosamine-6-sulfatase, Vol. 11, p. 267 1.2.3.14 abscisic-aldehyde oxidase, Vol. S1, p. 176 3.2.1.49 a-N-acetylgalactosaminidase, Vol. 13,p.10 1.2.1.10 acetaldehyde dehydrogenase (acetylating), Vol. 20, 3.2.1.53 b-N-acetylgalactosaminidase, Vol. 13,p.91 p. 115 2.4.99.3 a-N-acetylgalactosaminide a-2,6-sialyltransferase, 3.5.1.63 4-acetamidobutyrate deacetylase, Vol. 14,p.528 Vol. 33,p.335 3.5.1.51 4-acetamidobutyryl-CoA deacetylase, Vol. 14, 2.4.1.147 acetylgalactosaminyl-O-glycosyl-glycoprotein b- p. 482 1,3-N-acetylglucosaminyltransferase, Vol. 32, 3.5.1.29 2-(acetamidomethylene)succinate hydrolase, p. 287 Vol.