Generated by SRI International Pathway Tools Version 25.0, Authors S
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

An Atpase Domain Common to Prokaryotic Cell Cycle Proteins
Proc. Natl. Acad. Sci. USA Vol. 89, pp. 7290-7294, August 1992 Biochemistry An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp7O heat shock proteins (structural comparison/property pattern/remote homology) PEER BORK, CHRIS SANDER, AND ALFONSO VALENCIA European Molecular Biology Laboratory, D-6900 Heidelberg, Federal Republic of Germany Communicated by Russell F. Doolittle, March 6, 1992 ABSTRACT The functionally diverse actin, hexokinase, and hsp7O protein families have in common an ATPase domain of known three-dimensional structure. Optimal superposition ofthe three structures and alignment ofmany sequences in each of the three families has revealed a set of common conserved residues, distributed in five sequence motifs, which are in- volved in ATP binding and in a putative interdomain hinge. From the multiple sequence aliment in these motifs a pattern of amino acid properties required at each position is defined. The discriminatory power of the pattern is in part due to the use of several known three-dimensional structures and many sequences and in part to the "property" method ofgeneralizing from observed amino acid frequencies to amino acid fitness at each sequence position. A sequence data base search with the pattern significantly matches sugar kinases, such as fuco-, glucono-, xylulo-, ribulo-, and glycerokinase, as well as the prokaryotic cell cycle proteins MreB, FtsA, and StbA. These are predicted to have subdomains with the same tertiary structure as the ATPase subdomains Ia and Ha of hexokinase, actin, and Hsc7O, a very similar ATP binding pocket, and the capacity for interdomain hinge motion accompanying func- tional state changes. -

Review Article RNA Degradation in Staphylococcus Aureus: Diversity of Ribonucleases and Their Impact
Hindawi Publishing Corporation International Journal of Genomics Volume 2015, Article ID 395753, 12 pages http://dx.doi.org/10.1155/2015/395753 Review Article RNA Degradation in Staphylococcus aureus: Diversity of Ribonucleases and Their Impact Rémy A. Bonnin and Philippe Bouloc Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, UniversiteParis-Sud,Universit´ eParis-Saclay,91400Orsay,France´ Correspondence should be addressed to Remy´ A. Bonnin; [email protected] Received 6 January 2015; Accepted 4 March 2015 Academic Editor: Martine A. Collart Copyright © 2015 R. A. Bonnin and P. Bouloc. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The regulation of RNA decay is now widely recognized as having a central role in bacterial adaption to environmental stress.Here we present an overview on the diversity of ribonucleases (RNases) and their impact at the posttranscriptional level in the human pathogen Staphylococcus aureus. RNases in prokaryotes have been mainly studied in the two model organisms Escherichia coli and Bacillus subtilis. Based on identified RNases in these two models, putative orthologs have been identified in S. aureus.Themain staphylococcal RNases involved in the processing and degradation of the bulk RNA are (i) endonucleases RNase III and RNase Y and (ii) exonucleases RNase J1/J2 and PNPase, having 5 to 3 and 3 to 5 activities, respectively. The diversity and potential roles of each RNase and of Hfq and RppH are discussed in the context of recent studies, some of which are based on next-generation sequencing technology. -

Generated by SRI International Pathway Tools Version 25.0, Authors S
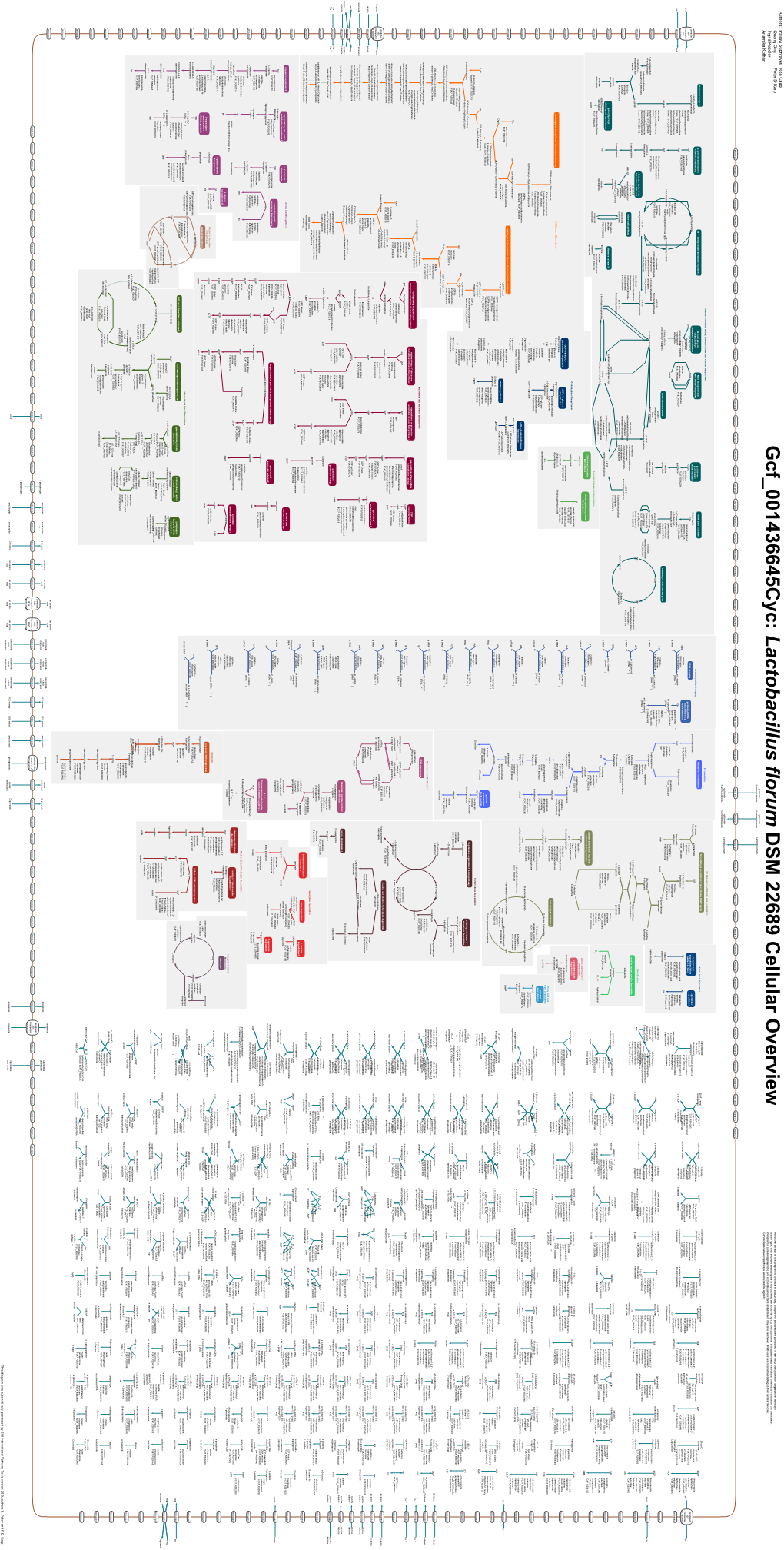
An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000238675-HmpCyc: Bacillus smithii 7_3_47FAA Cellular Overview Connections between pathways are omitted for legibility. -

Polymerase Ribozyme with Promoter Recognition
In vitro Evolution of a Processive Clamping RNA Polymerase Ribozyme with Promoter Recognition by Razvan Cojocaru BSc, Simon Fraser University, 2014 Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in the Department of Molecular Biology and Biochemistry Faculty of Science © Razvan Cojocaru 2021 SIMON FRASER UNIVERSITY Summer 2021 Copyright in this work is held by the author. Please ensure that any reproduction or re-use is done in accordance with the relevant national copyright legislation. Declaration of Committee Name: Razvan Cojocaru Degree: Doctor of Philosophy Title: In vitro Evolution of a Processive Clamping RNA Polymerase Ribozyme with Promoter Recognition Committee: Chair: Lisa Craig Professor, Molecular Biology and Biochemistry Peter Unrau Supervisor Professor, Molecular Biology and Biochemistry Dipankar Sen Committee Member Professor, Molecular Biology and Biochemistry Michel Leroux Committee Member Professor, Molecular Biology and Biochemistry Mani Larijani Internal Examiner Associate Professor, Molecular Biology and Biochemistry Gerald Joyce External Examiner Professor, Jack H. Skirball Center for Chemical Biology and Proteomics Salk Institute for Biological Studies Date Defended/Approved: August 12, 2021 ii Abstract The RNA World hypothesis proposes that the early evolution of life began with RNAs that can serve both as carriers of genetic information and as catalysts. Later in evolution, these functions were gradually replaced by DNA and enzymatic proteins in cellular biology. I start by reviewing the naturally occurring catalytic RNAs, ribozymes, as they play many important roles in biology today. These ribozymes are central to protein synthesis and the regulation of gene expression, creating a landscape that strongly supports an early RNA World. -

| Hai Lui a Un Acutul Luniit Moonhiti
|HAI LUI AUN ACUTULUS010006055B2 LUNIIT MOONHITI (12 ) United States Patent (10 ) Patent No. : US 10 , 006 , 055 B2 Burk et al. (45 ) Date of Patent: Jun . 26 , 2018 ( 54 ) MICROORGANISMS FOR PRODUCING 2002/ 0168654 A1 11/ 2002 Maranas et al. 2003 / 0059792 Al 3 /2003 Palsson et al . BUTADIENE AND METHODS RELATED 2003 /0087381 A1 5 / 2003 Gokarn THERETO 2003 / 0224363 Al 12 /2003 Park et al . 2003 / 0233218 Al 12 /2003 Schilling (71 ) Applicant: Genomatica , Inc. , San Diego , CA (US ) 2004 / 0009466 AL 1 /2004 Maranas et al. 2004 / 0029149 Al 2 /2004 Palsson et al. ( 72 ) Inventors : Mark J . Burk , San Diego , CA (US ) ; 2004 / 0072723 A1 4 /2004 Palsson et al. Anthony P . Burgard , Bellefonte , PA 2004 / 0152159 Al 8 / 2004 Causey et al . 2005 /0042736 A1 2 / 2005 San et al . (US ) ; Robin E . Osterhout , San Diego , 2005 / 0079482 A1 4 / 2005 Maranas et al . CA (US ) ; Jun Sun , San Diego , CA 2006 / 0046288 Al 3 / 2006 Ka - Yiu et al. ( US ) ; Priti Pharkya , San Diego , CA 2006 / 0073577 A1 4 / 2006 Ka - Yiu et al . (US ) 2007 /0184539 Al 8 / 2007 San et al . 2009 / 0047718 Al 2 / 2009 Blaschek et al . 2009 / 0047719 Al 2 / 2009 Burgard et al . (73 ) Assignee : Genomatica , Inc ., San Diego , CA (US ) 2009 /0191593 A1 7 / 2009 Burk et al . 2010 / 0003716 A1 1 / 2010 Cervin et al. ( * ) Notice : Subject to any disclaimer , the term of this 2010 /0184171 Al 7 /2010 Jantama et al. patent is extended or adjusted under 35 2010 /0304453 Al 12 / 2010 Trawick et al . -

Supplementary Materials
Supplementary Materials Figure S1. Differentially abundant spots between the mid-log phase cells grown on xylan or xylose. Red and blue circles denote spots with increased and decreased abundance respectively in the xylan growth condition. The identities of the circled spots are summarized in Table 3. Figure S2. Differentially abundant spots between the stationary phase cells grown on xylan or xylose. Red and blue circles denote spots with increased and decreased abundance respectively in the xylan growth condition. The identities of the circled spots are summarized in Table 4. S2 Table S1. Summary of the non-polysaccharide degrading proteins identified in the B. proteoclasticus cytosol by 2DE/MALDI-TOF. Protein Locus Location Score pI kDa Pep. Cov. Amino Acid Biosynthesis Acetylornithine aminotransferase, ArgD Bpr_I1809 C 1.7 × 10−4 5.1 43.9 11 34% Aspartate/tyrosine/aromatic aminotransferase Bpr_I2631 C 3.0 × 10−14 4.7 43.8 15 46% Aspartate-semialdehyde dehydrogenase, Asd Bpr_I1664 C 7.6 × 10−18 5.5 40.1 17 50% Branched-chain amino acid aminotransferase, IlvE Bpr_I1650 C 2.4 × 10−12 5.2 39.2 13 32% Cysteine synthase, CysK Bpr_I1089 C 1.9 × 10−13 5.0 32.3 18 72% Diaminopimelate dehydrogenase Bpr_I0298 C 9.6 × 10−16 5.6 35.8 16 49% Dihydrodipicolinate reductase, DapB Bpr_I2453 C 2.7 × 10−6 4.9 27.0 9 46% Glu/Leu/Phe/Val dehydrogenase Bpr_I2129 C 1.2 × 10−30 5.4 48.6 31 64% Imidazole glycerol phosphate synthase Bpr_I1240 C 8.0 × 10−3 4.7 22.5 8 44% glutamine amidotransferase subunit Ketol-acid reductoisomerase, IlvC Bpr_I1657 C 3.8 × 10−16 -

(12) Patent Application Publication (10) Pub. No.: US 2014/0155567 A1 Burk Et Al
US 2014O155567A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2014/0155567 A1 Burk et al. (43) Pub. Date: Jun. 5, 2014 (54) MICROORGANISMS AND METHODS FOR (60) Provisional application No. 61/331,812, filed on May THE BIOSYNTHESIS OF BUTADENE 5, 2010. (71) Applicant: Genomatica, Inc., San Diego, CA (US) Publication Classification (72) Inventors: Mark J. Burk, San Diego, CA (US); (51) Int. Cl. Anthony P. Burgard, Bellefonte, PA CI2P 5/02 (2006.01) (US); Jun Sun, San Diego, CA (US); CSF 36/06 (2006.01) Robin E. Osterhout, San Diego, CA CD7C II/6 (2006.01) (US); Priti Pharkya, San Diego, CA (52) U.S. Cl. (US) CPC ................. CI2P5/026 (2013.01); C07C II/I6 (2013.01); C08F 136/06 (2013.01) (73) Assignee: Genomatica, Inc., San Diego, CA (US) USPC ... 526/335; 435/252.3:435/167; 435/254.2: (21) Appl. No.: 14/059,131 435/254.11: 435/252.33: 435/254.21:585/16 (22) Filed: Oct. 21, 2013 (57) ABSTRACT O O The invention provides non-naturally occurring microbial Related U.S. Application Data organisms having a butadiene pathway. The invention addi (63) Continuation of application No. 13/101,046, filed on tionally provides methods of using Such organisms to produce May 4, 2011, now Pat. No. 8,580,543. butadiene. Patent Application Publication Jun. 5, 2014 Sheet 1 of 4 US 2014/O155567 A1 ?ueudos!SMS |?un61– Patent Application Publication Jun. 5, 2014 Sheet 2 of 4 US 2014/O155567 A1 VOJ OO O Z?un61– Patent Application Publication US 2014/O155567 A1 {}}} Hººso Patent Application Publication Jun. -
Generate Metabolic Map Poster
Authors: Peter D. Karp Suzanne Paley Julio Collado-Vides John L Ingraham Ingrid Keseler Markus Krummenacker Cesar Bonavides-Martinez Robert Gunsalus An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Carol Fulcher Ian Paulsen Socorro Gama-Castro Robert LaRossa Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. EcoCyc: Escherichia coli K-12 substr. MG1655 Cellular Overview Connections between pathways are omitted for legibility. Anamika Kothari Amanda Mackie Alberto Santos-Zavaleta succinate phosphate succinate N-acetyl-DL-methionine + L-ornithine glutathione + L-methionine S-oxide D-fructofuranose γ Ag+ molybdate ferroheme b L,L-homocystine asp lys cys L-alanyl- -D- D-mannopyranose 6-phosphate 2+ 2+ H D-methionine 2-deoxy-D-glucose succinate formate formate succinate D-tartrate putrescine agmatine cadaverine L-tartrate D-fructofuranose 6-phosphate + nitrate nitrate Cu thiosulfate deoxycholate L,L-homocystine D-cystine D-cycloserine methyl β-D-glucoside putrescine asp spermidine (S)-2-hydroxybutanoate (S)-2-hydroxybutanoate arg L-homoserine lactone magnesium hydrogenphosphate magnesium hydrogenphosphate antimonous acid glutamyl-meso- Co2+ Cd2+ lactulose poly-β-1,6- met cob(I)inamide 2,3-dioxo- -

Table S1. List of Oligonucleotide Primers Used
Table S1. List of oligonucleotide primers used. Cla4 LF-5' GTAGGATCCGCTCTGTCAAGCCTCCGACC M629Arev CCTCCCTCCATGTACTCcgcGATGACCCAgAGCTCGTTG M629Afwd CAACGAGCTcTGGGTCATCgcgGAGTACATGGAGGGAGG LF-3' GTAGGCCATCTAGGCCGCAATCTCGTCAAGTAAAGTCG RF-5' GTAGGCCTGAGTGGCCCGAGATTGCAACGTGTAACC RF-3' GTAGGATCCCGTACGCTGCGATCGCTTGC Ukc1 LF-5' GCAATATTATGTCTACTTTGAGCG M398Arev CCGCCGGGCAAgAAtTCcgcGAGAAGGTACAGATACGc M398Afwd gCGTATCTGTACCTTCTCgcgGAaTTcTTGCCCGGCGG LF-3' GAGGCCATCTAGGCCATTTACGATGGCAGACAAAGG RF-5' GTGGCCTGAGTGGCCATTGGTTTGGGCGAATGGC RF-3' GCAATATTCGTACGTCAACAGCGCG Nrc2 LF-5' GCAATATTTCGAAAAGGGTCGTTCC M454Grev GCCACCCATGCAGTAcTCgccGCAGAGGTAGAGGTAATC M454Gfwd GATTACCTCTACCTCTGCggcGAgTACTGCATGGGTGGC LF-3' GAGGCCATCTAGGCCGACGAGTGAAGCTTTCGAGCG RF-5' GAGGCCTGAGTGGCCTAAGCATCTTGGCTTCTGC RF-3' GCAATATTCGGTCAACGCTTTTCAGATACC Ipl1 LF-5' GTCAATATTCTACTTTGTGAAGACGCTGC M629Arev GCTCCCCACGACCAGCgAATTCGATagcGAGGAAGACTCGGCCCTCATC M629Afwd GATGAGGGCCGAGTCTTCCTCgctATCGAATTcGCTGGTCGTGGGGAGC LF-3' TGAGGCCATCTAGGCCGGTGCCTTAGATTCCGTATAGC RF-5' CATGGCCTGAGTGGCCGATTCTTCTTCTGTCATCGAC RF-3' GACAATATTGCTGACCTTGTCTACTTGG Ire1 LF-5' GCAATATTAAAGCACAACTCAACGC D1014Arev CCGTAGCCAAGCACCTCGgCCGAtATcGTGAGCGAAG D1014Afwd CTTCGCTCACgATaTCGGcCGAGGTGCTTGGCTACGG LF-3' GAGGCCATCTAGGCCAACTGGGCAAAGGAGATGGA RF-5' GAGGCCTGAGTGGCCGTGCGCCTGTGTATCTCTTTG RF-3' GCAATATTGGCCATCTGAGGGCTGAC Kin28 LF-5' GACAATATTCATCTTTCACCCTTCCAAAG L94Arev TGATGAGTGCTTCTAGATTGGTGTCggcGAAcTCgAGCACCAGGTTG L94Afwd CAACCTGGTGCTcGAgTTCgccGACACCAATCTAGAAGCACTCATCA LF-3' TGAGGCCATCTAGGCCCACAGAGATCCGCTTTAATGC RF-5' CATGGCCTGAGTGGCCAGGGCTAGTACGACCTCG -
When the Reaction Is
Table S3. iJL1678-ME model modification (blocked reactions) Iter. Cat. ID Name Formula Subsystem Comments (When the reaction is turned on) 1 bp2 EDD 6-phosphogluconate dehydratase 6pgc_c⇌2ddg6p_c + h2o_c Pentose Phosphate Pathway Create a major effect of steep acetate overflow elevation in high growth. Comparing to the main glycolytic pathway, it is metabolicly less efficient but proteomicly more efficient. bp1 ICL Isocitrate lyase icit_c→glx_c + succ_c Anaplerotic Reactions Bypass for the main TCA cycle pathways from turning isocitrate to succinate, when ICL is turned on, Isocitrate dehydrogenase(ICDHyr), 2-Oxogluterate dehydrogenase(AKGDH) and Succinyl-CoA synthetase (ATP-forming,SUCOAS) would reduce. Ref. (1) and (2) shows that this reaction is off in higher growth. Ref. (3) shows that this reaction is converging to being off when the dynamic of respiration using enzyme kinetics is simulated. 2 bp1 ABTA 4-aminobutyrate transaminase 4abut_c + akg_c⇌glu__L_c + sucsal_c Arginine and Proline Metabolism Another backup pathway of succinate production, from 2-Oxoglutarate (akg). Respiration would be induced when it is on, since the flux through ETC(CYTBO3_4pp and ATPS4rpp) would increase. As it requires the co-factor pyridoxal 5'-phosphate(2−) to get catalyzed(4), indicating that this reaction is regulated by the flux of other reactions(pyridoxal 5'- phosphate(2-) production, etc.). 3 GLYAT Glycine C-acetyltransferase accoa_c + gly_c⇌2aobut_c + coa_c Glycine and Serine Metabolism A reaction that back up for the respiration. Reactions fluxes in TCA cycle would drop when this reaction is turned on. It also requires pyridoxal 5'-phosphate(2−) for the regulation. 4 NADTRHD NAD transhydrogenase nad_c + nadph_c⇌nadh_c + nadp_c Oxidative Phosphorylation A reaction that would make the transition between NAD and NADP metabolically more efficient. -

(12) Patent Application Publication (10) Pub. No.: US 2010/0317005 A1 Hardin Et Al
US 20100317005A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2010/0317005 A1 Hardin et al. (43) Pub. Date: Dec. 16, 2010 (54) MODIFIED NUCLEOTIDES AND METHODS (22) Filed: Mar. 15, 2010 FOR MAKING AND USE SAME Related U.S. Application Data (63) Continuation of application No. 11/007,794, filed on Dec. 8, 2004, now abandoned, which is a continuation (75) Inventors: Susan H. Hardin, College Station, in-part of application No. 09/901,782, filed on Jul. 9, TX (US); Hongyi Wang, Pearland, 2001. TX (US); Brent A. Mulder, (60) Provisional application No. 60/527,909, filed on Dec. Sugarland, TX (US); Nathan K. 8, 2003, provisional application No. 60/216,594, filed Agnew, Richmond, TX (US); on Jul. 7, 2000. Tommie L. Lincecum, JR., Publication Classification Houston, TX (US) (51) Int. Cl. CI2O I/68 (2006.01) Correspondence Address: (52) U.S. Cl. ............................................................ 435/6 LIFE TECHNOLOGES CORPORATION (57) ABSTRACT CFO INTELLEVATE Labeled nucleotide triphosphates are disclosed having a label P.O. BOX S2OSO bonded to the gamma phosphate of the nucleotide triphos MINNEAPOLIS, MN 55402 (US) phate. Methods for using the gamma phosphate labeled nucleotide are also disclosed where the gamma phosphate labeled nucleotide are used to attach the labeled gamma phos (73) Assignees: LIFE TECHNOLOGIES phate in a catalyzed (enzyme or man-made catalyst) reaction to a target biomolecule or to exchange a phosphate on a target CORPORATION, Carlsbad, CA biomolecule with a labeled gamme phosphate. Preferred tar (US); VISIGEN get biomolecules are DNAs, RNAs, DNA/RNAs, PNA, BIOTECHNOLOGIES, INC. polypeptide (e.g., proteins enzymes, protein, assemblages, etc.), Sugars and polysaccharides or mixed biomolecules hav ing two or more of DNAs, RNAs, DNA/RNAs, polypeptide, (21) Appl. -

Characterization of Human UMP/CMP Kinase and Its Phosphorylation of D- and 1 L-Form Deoxycytidine Analogue Monophosphates
[CANCER RESEARCH 62, 1624–1631, March 15, 2002] Characterization of Human UMP/CMP Kinase and Its Phosphorylation of D- and 1 L-Form Deoxycytidine Analogue Monophosphates Jieh-Yuan Liou, Ginger E. Dutschman, Wing Lam, Zaoli Jiang, and Yung-Chi Cheng2 Department of Pharmacology, Yale University School of Medicine, New Haven, Connecticut 06520 ABSTRACT with leukemia, lymphoma, or solid tumors (11). Deoxycytidine ana- logues, such as -D-2Ј,3Ј-dideoxycytidine and L-(Ϫ)-SddC (Lamivu- Pyrimidine nucleoside monophosphate kinase [UMP/CMP kinase dine), have been shown to have anti-HIV and antihuman hepatitis B (UMP/CMPK); EC 2.7.4.14] plays a crucial role in the formation of UDP, virus activities (12–17). L-(Ϫ)-SddC was the first nucleoside analogue CDP, and dCDP, which are required for cellular nucleic acid synthesis. Several cytidine and deoxycytidine analogues are important anticancer with an L configuration to show therapeutic activity and, thus, defined  Ј Ј and antiviral drugs. These drugs require stepwise phosphorylation to their a new category for the design of nucleoside analogues. -L-2 ,3 - triphosphate forms to exert their therapeutic effects. The role of UMP/ dideoxy-5-fluoro-3Ј-thia-cytidine and -L-2Ј,3Ј-dideoxy-2Ј,3Ј-dide- CMPK for the phosphorylation of nucleoside analogues has been indi- hydro-5-fluorocytidine have been shown to be potent antihuman hep- cated. Thus, we cloned the human UMP/CMPK gene, expressed it in atitis B virus agents in vitro and in animal studies (18–22). In studies Escherichia coli, and purified it to homogeneity. Its kinetic properties of other -L-(Ϫ)-2Ј,3Ј-dideoxycytidine analogues, it was observed were determined.