Structures of B. Subtilis Maturation Rnases Captured on 50S
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Review Article RNA Degradation in Staphylococcus Aureus: Diversity of Ribonucleases and Their Impact
Hindawi Publishing Corporation International Journal of Genomics Volume 2015, Article ID 395753, 12 pages http://dx.doi.org/10.1155/2015/395753 Review Article RNA Degradation in Staphylococcus aureus: Diversity of Ribonucleases and Their Impact Rémy A. Bonnin and Philippe Bouloc Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, UniversiteParis-Sud,Universit´ eParis-Saclay,91400Orsay,France´ Correspondence should be addressed to Remy´ A. Bonnin; [email protected] Received 6 January 2015; Accepted 4 March 2015 Academic Editor: Martine A. Collart Copyright © 2015 R. A. Bonnin and P. Bouloc. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The regulation of RNA decay is now widely recognized as having a central role in bacterial adaption to environmental stress.Here we present an overview on the diversity of ribonucleases (RNases) and their impact at the posttranscriptional level in the human pathogen Staphylococcus aureus. RNases in prokaryotes have been mainly studied in the two model organisms Escherichia coli and Bacillus subtilis. Based on identified RNases in these two models, putative orthologs have been identified in S. aureus.Themain staphylococcal RNases involved in the processing and degradation of the bulk RNA are (i) endonucleases RNase III and RNase Y and (ii) exonucleases RNase J1/J2 and PNPase, having 5 to 3 and 3 to 5 activities, respectively. The diversity and potential roles of each RNase and of Hfq and RppH are discussed in the context of recent studies, some of which are based on next-generation sequencing technology. -

Generated by SRI International Pathway Tools Version 25.0, Authors S
An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000238675-HmpCyc: Bacillus smithii 7_3_47FAA Cellular Overview Connections between pathways are omitted for legibility. -
When the Reaction Is
Table S3. iJL1678-ME model modification (blocked reactions) Iter. Cat. ID Name Formula Subsystem Comments (When the reaction is turned on) 1 bp2 EDD 6-phosphogluconate dehydratase 6pgc_c⇌2ddg6p_c + h2o_c Pentose Phosphate Pathway Create a major effect of steep acetate overflow elevation in high growth. Comparing to the main glycolytic pathway, it is metabolicly less efficient but proteomicly more efficient. bp1 ICL Isocitrate lyase icit_c→glx_c + succ_c Anaplerotic Reactions Bypass for the main TCA cycle pathways from turning isocitrate to succinate, when ICL is turned on, Isocitrate dehydrogenase(ICDHyr), 2-Oxogluterate dehydrogenase(AKGDH) and Succinyl-CoA synthetase (ATP-forming,SUCOAS) would reduce. Ref. (1) and (2) shows that this reaction is off in higher growth. Ref. (3) shows that this reaction is converging to being off when the dynamic of respiration using enzyme kinetics is simulated. 2 bp1 ABTA 4-aminobutyrate transaminase 4abut_c + akg_c⇌glu__L_c + sucsal_c Arginine and Proline Metabolism Another backup pathway of succinate production, from 2-Oxoglutarate (akg). Respiration would be induced when it is on, since the flux through ETC(CYTBO3_4pp and ATPS4rpp) would increase. As it requires the co-factor pyridoxal 5'-phosphate(2−) to get catalyzed(4), indicating that this reaction is regulated by the flux of other reactions(pyridoxal 5'- phosphate(2-) production, etc.). 3 GLYAT Glycine C-acetyltransferase accoa_c + gly_c⇌2aobut_c + coa_c Glycine and Serine Metabolism A reaction that back up for the respiration. Reactions fluxes in TCA cycle would drop when this reaction is turned on. It also requires pyridoxal 5'-phosphate(2−) for the regulation. 4 NADTRHD NAD transhydrogenase nad_c + nadph_c⇌nadh_c + nadp_c Oxidative Phosphorylation A reaction that would make the transition between NAD and NADP metabolically more efficient. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Genome-Wide Investigation of Cellular Functions for Trna Nucleus
Genome-wide Investigation of Cellular Functions for tRNA Nucleus- Cytoplasm Trafficking in the Yeast Saccharomyces cerevisiae DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Hui-Yi Chu Graduate Program in Molecular, Cellular and Developmental Biology The Ohio State University 2012 Dissertation Committee: Anita K. Hopper, Advisor Stephen Osmani Kurt Fredrick Jane Jackman Copyright by Hui-Yi Chu 2012 Abstract In eukaryotic cells tRNAs are transcribed in the nucleus and exported to the cytoplasm for their essential role in protein synthesis. This export event was thought to be unidirectional. Surprisingly, several lines of evidence showed that mature cytoplasmic tRNAs shuttle between nucleus and cytoplasm and their distribution is nutrient-dependent. This newly discovered tRNA retrograde process is conserved from yeast to vertebrates. Although how exactly the tRNA nuclear-cytoplasmic trafficking is regulated is still under investigation, previous studies identified several transporters involved in tRNA subcellular dynamics. At least three members of the β-importin family function in tRNA nuclear-cytoplasmic intracellular movement: (1) Los1 functions in both the tRNA primary export and re-export processes; (2) Mtr10, directly or indirectly, is responsible for the constitutive retrograde import of cytoplasmic tRNA to the nucleus; (3) Msn5 functions solely in the re-export process. In this thesis I focus on the physiological role(s) of the tRNA nuclear retrograde pathway. One possibility is that nuclear accumulation of cytoplasmic tRNA serves to modulate translation of particular transcripts. To test this hypothesis, I compared expression profiles from non-translating mRNAs and polyribosome-bound translating mRNAs collected from msn5Δ and mtr10Δ mutants and wild-type cells, in fed or acute amino acid starvation conditions. -

FUNCTIONAL and MECHANISTIC CHARACTERIZATION of TWO TRNA MODIFYING ENZYMES LAURA CAROLE KEFFER-WILKES Master of Science, Universi
FUNCTIONAL AND MECHANISTIC CHARACTERIZATION OF TWO TRNA MODIFYING ENZYMES LAURA CAROLE KEFFER-WILKES Master of Science, University of Lethbridge, 2012 A Thesis Submitted to the School of Graduate Studies Of the University of Lethbridge In Partial Fulfillment of the Requirements for the Degree DOCTOR OF PHILOSOPHY Department of Chemistry and Biochemistry University of Lethbridge LETHBRIDGE, ALBERTA, CANADA © Laura Keffer-Wilkes, 2016 FUNCTIONAL AND MECHANISTIC CHARACTERIZATION OF TWO TRNA MODIFYING ENZYMES LAURA CAROLE KEFFER-WILKES Date of Defence: June 28, 2016 Dr. Ute Wieden-Kothe Associate Professor Ph.D. Supervisor Dr. Tony Russell Assistant Professor Ph.D. Thesis Examination Committee Member Dr. Stacey Wetmore Professor Ph.D. Thesis Examination Committee Member Dr. Hans-Joachim Wieden Professor Ph.D. Thesis Examination Committee Member Dr. Eugene Mueller Professor Ph.D. External Examiner University of Louisville Louisville, Kentucky Dr. Elizabeth Schultz Associate Professor Ph.D. Internal External Examiner University of Lethbridge Dr. Michael Gerken Professor Ph.D. Chair, Thesis Examination Committee Abstract The formation of pseudouridine (Ѱ) and 5-methyluridine (m5U) in the T-arm of transfer RNAs (tRNAs) is near-universally conserved. These two modifications are formed in Escherichia coli by the pseudouridine synthase TruB and the S-adenosylmethionine- dependent methyltransferase TrmA, respectively. In this thesis, I investigate the function and mechanisms of these two tRNA modifying enzymes. First, in vitro and in vivo analysis of TruB reveals that this enzyme is acting as a tRNA chaperone which proves a long outstanding hypothesis. Secondly, characterization of ligand binding by TrmA shows that binding is cooperative and disruption of tRNA elbow region tertiary interactions by TrmA is essential for efficient tRNA binding and catalysis, leading to future analysis. -

A Protein Subunit of Human Rnase P, Rpp14, and Its Interacting Partner, OIP2, Have 335 Exoribonuclease Activity
A protein subunit of human RNase P, Rpp14, and its interacting partner, OIP2, have 335 exoribonuclease activity Taijiao Jiang and Sidney Altman* Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06520 Contributed by Sidney Altman, February 12, 2002 -The processing of precursor tRNAs at their 5 and 3 termini is a presence of [␣-32P]UTP. Internally labeled SupS1-3Ј was pre fundamental event in the biosynthesis of tRNA. RNase P is gener- pared from internally labeled 5Ј-SupS1-3Ј by removing the 5Ј ally responsible for endonucleolytic removal of a leader sequence leader sequence by using purified human RNase P. For the of precursor tRNA to generate the mature 5 terminus. However, synthesis of 5Ј-32P-labeled 5Ј-Tyr-3Ј, transcription reactions were much less is known about the RNase P counterparts or other performed with nonlabeled NTPs, the RNA was purified and proteins that are active at the tRNA 3 terminus. Here we show that then was treated with calf intestinal phosphatase, extracted two one of the human RNase P subunits, Rpp14, together with one of times with phenol͞chloroform and one time with chloroform. its interacting protein partners, OIP2, is a 335 exoribonuclease Recovery was by ethanol precipitation. 5Ј-Tyr-3Ј was labeled .with a phosphorolytic activity that processes the 3 terminus of with [␥-32P]ATP by using polynucleotide kinase precursor tRNA. Immunoprecipitates of a crude human RNase P Adult enhancer factor-1 (AEF) RNA, a single-stranded RNA complex can process both ends of precursor tRNA by hydrolysis, from Drosophila melanogaster (12), was labeled as indicated by but purified RNase P has no exonuclease activity. -

B Number Gene Name Strand Orientation Protein Length Mrna
list list sample) short list predicted B number Operon ID Gene name assignment Protein length mRNA present mRNA intensity Gene description Protein detected - Strand orientation Membrane protein detected (total list) detected (long list) membrane sample Proteins detected - detected (short list) # of tryptic peptides # of tryptic peptides # of tryptic peptides # of tryptic peptides # of tryptic peptides Functional category detected (membrane Protein detected - total Protein detected - long b0001 thrL + 21 1344 P 1 0 0 0 0 thr operon leader peptide Metabolism of small molecules 1 b0002 thrA + 820 13624 P 39 P 18 P 18 P 18 P(m) 2 aspartokinase I, homoserine dehydrogenase I Metabolism of small molecules 1 b0003 thrB + 310 6781 P 9 P 3 3 P 3 0 homoserine kinase Metabolism of small molecules 1 b0004 thrC + 428 15039 P 18 P 10 P 11 P 10 0 threonine synthase Metabolism of small molecules 1 b0005 b0005 + 98 432 A 5 0 0 0 0 orf, hypothetical protein Open reading frames 2 b0006 yaaA - 258 1047 P 11 P 1 2 P 1 0 orf, hypothetical protein Open reading frames 3 b0007 yaaJ - 476 342 P 8 0 0 0 0 MP-GenProt-PHD inner membrane transport protein Miscellaneous 4 b0008 talB + 317 20561 P 20 P 13 P 16 P 13 0 transaldolase B Metabolism of small molecules 5 b0009 mog + 195 1296 P 7 0 0 0 0 required for the efficient incorporation of molybdate into molybdoproteins Metabolism of small molecules 6 b0010 yaaH - 188 407 A 2 0 0 0 0 PHD orf, hypothetical protein Open reading frames 7 b0011 b0011 - 237 338 P 13 0 0 0 0 putative oxidoreductase Miscellaneous 8 b0012 htgA -

A Possible Functional Link Between RNA Degradation and Transcription in Bacillus Subtilis
A possible functional link between RNA degradation and transcription in Bacillus subtilis Dissertation for the award of the degree “Doctor of Philosophy” Division of Mathematics and Natural Sciences of the Georg-August-University Göttingen within the program “Microbiology and Biochemistry” of the Georg-August University School of Science (GAUSS) submitted by Martin Benda from Prague (Czech Republic) Göttingen 2020 Thesis Committee Prof. Dr. Jörg Stülke (Supervisor and 1st Reviewer) Institute for Microbiology and Genetics, Department of General Microbiology, Georg-August-University Göttingen Prof. Dr. Rolf Daniel (2nd Reviewer) Institute for Microbiology and Genetics, Department of Genomic and Applied Microbiology, Georg-August- University Göttingen Prof. Dr. Fabian M. Commichau Institute for Biotechnology, Department of Synthetic Microbiology, Brandenburg University of Technology Cottbus-Senftenberg Additional members of the Examination Board Prof. Dr. Markus Bohnsack Department of Molecular Biology, University Medical Center Göttingen Prof. Dr. Patrick Cramer Department of Molecular Biology, Max Planck Institute for Biophysical Chemistry, Göttingen Prof. Dr. Stefanie Pöggeler Institute for Microbiology and Genetics, Department of Genetics of Eukaryotic Microorganisms, Georg- August-University Göttingen Date of oral examination: September 17th, 2020 I hereby declare that this Ph.D. thesis entitled “A possible functional link between RNA degradation and transcription in Bacillus subtilis” has been written independently and with no other sources and aids than quoted. Martin Benda Acknowledgements First of all, I would like to express my deep thanks to my supervisor Prof. Dr. Jörg Stülke for the great guidance during this work. He put his full trust in me and gave me the opportunity to work on this intricate, but engaging topic of RNA degradation. -

An Investigation of Bacterial Ribonucleases As an Antibiotic Target Ashley Denise Frazier East Tennessee State University
East Tennessee State University Digital Commons @ East Tennessee State University Electronic Theses and Dissertations Student Works 5-2012 An Investigation of Bacterial Ribonucleases as an Antibiotic Target Ashley Denise Frazier East Tennessee State University Follow this and additional works at: https://dc.etsu.edu/etd Part of the Bacteriology Commons Recommended Citation Frazier, Ashley Denise, "An Investigation of Bacterial Ribonucleases as an Antibiotic Target" (2012). Electronic Theses and Dissertations. Paper 1417. https://dc.etsu.edu/etd/1417 This Dissertation - Open Access is brought to you for free and open access by the Student Works at Digital Commons @ East Tennessee State University. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of Digital Commons @ East Tennessee State University. For more information, please contact [email protected]. An Investigation of Bacterial Ribonucleases as an Antibiotic Target A dissertation presented to the faculty of the Department of Biochemistry and Molecular Biology East Tennessee State University In partial fulfillment of the requirements for the degree Doctor of Philosophy of Biomedical Sciences by Ashley Denise Frazier May 2012 W. Scott Champney, Ph.D., Chair Mitchell E. Robinson, Ph.D. Douglas P. Thewke, Ph.D. J. Russell Hayman, Ph.D. Bert C. Lampson, Ph.D. Keywords: Ribonuclease Mutants, Ribosomal Subunit Assembly, Aminoglycosides, Escherichia coli, Staphylococcus aureus, Protein Synthesis, Vanadyl Ribonucleoside Complex ABSTRACT An Investigation of Bacterial Ribonucleases as an Antibiotic Target by Ashley Denise Frazier Antibiotics have been commonly used in medical practice for over 40 years. However, the misuse and overuse of current antibiotics is thought to be the primary cause for the increase in antibiotic resistance. -

Tese Jucimar Zacaria.Pdf (4.725Mb)
UNIVERSIDADE DE CAXIAS DO SUL CENTRO DE CIÊNCIAS BIOLÓGICAS E DA SAÚDE INSTITUTO DE BIOTECNOLOGIA PROGRAMA DE PÓS-GRADUAÇÃO EM BIOTECNOLOGIA DIVERSIDADE, CLONAGEM E CARACTERIZAÇÃO DE NUCLEASES EXTRACELULARES DE Aeromonas spp. JUCIMAR ZACARIA CAXIAS DO SUL 2016 JUCIMAR ZACARIA DIVERSIDADE, CLONAGEM E CARACTERIZAÇÃO DE NUCLEASES EXTRACELULARES DE Aeromonas spp. Tese apresentada ao programa de Pós- graduação em Biotecnologia da Universidade de Caxias do Sul, visando à obtenção de grau de Doutor em Biotecnologia. Orientador: Dr. Sergio Echeverrigaray Co-orientador: Dra. Ana Paula Longaray Delamare Caxias do Sul 2016 ii Z13d Zacaria, Jucimar Diversidade, clonagem e caracterização de nucleases extracelulares de Aeromonas spp. / Jucimar Zacaria. – 2016. 258 f.: il. Tese (Doutorado) - Universidade de Caxias do Sul, Programa de Pós- Graduação em Biotecnologia, 2016. Orientação: Sergio Echeverrigaray. Coorientação: Ana Paula Longaray Delamare. 1. Aeromas. 2. DNases extracelulares. 3. Termoestabilidade. 4. Dns. 5. Aha3441. I. Echeverrigaray, Sergio, orient. II. Delamare, Ana Paula Longaray, coorient. III. Título. Elaborado pelo Sistema de Geração Automática da UCS com os dados fornecidos pelo(a) autor(a). JUCIMAR ZACARIA DIVERSIDADE, CLONAGEM E CARACTERIZAÇÃO DE NUCLEASES EXTRACELULARES DE Aeromonas spp. Tese apresentada ao Programa de Pós-graduação em Biotecnologia da Universidade de Caxias do Sul, visando à obtenção do título de Doutor em Biotecnologia. Orientador: Prof. Dr. Sergio Echeverrigaray Laguna Co-orientadora: Profa. Dra. Ana Paula Longaray -

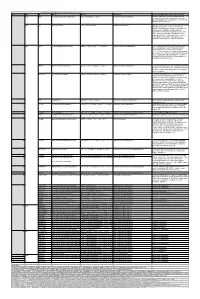
Supplemental Table S1: Comparison of the Deleted Genes in the Genome-Reduced Strains
Supplemental Table S1: Comparison of the deleted genes in the genome-reduced strains Legend 1 Locus tag according to the reference genome sequence of B. subtilis 168 (NC_000964) Genes highlighted in blue have been deleted from the respective strains Genes highlighted in green have been inserted into the indicated strain, they are present in all following strains Regions highlighted in red could not be deleted as a unit Regions highlighted in orange were not deleted in the genome-reduced strains since their deletion resulted in severe growth defects Gene BSU_number 1 Function ∆6 IIG-Bs27-47-24 PG10 PS38 dnaA BSU00010 replication initiation protein dnaN BSU00020 DNA polymerase III (beta subunit), beta clamp yaaA BSU00030 unknown recF BSU00040 repair, recombination remB BSU00050 involved in the activation of biofilm matrix biosynthetic operons gyrB BSU00060 DNA-Gyrase (subunit B) gyrA BSU00070 DNA-Gyrase (subunit A) rrnO-16S- trnO-Ala- trnO-Ile- rrnO-23S- rrnO-5S yaaC BSU00080 unknown guaB BSU00090 IMP dehydrogenase dacA BSU00100 penicillin-binding protein 5*, D-alanyl-D-alanine carboxypeptidase pdxS BSU00110 pyridoxal-5'-phosphate synthase (synthase domain) pdxT BSU00120 pyridoxal-5'-phosphate synthase (glutaminase domain) serS BSU00130 seryl-tRNA-synthetase trnSL-Ser1 dck BSU00140 deoxyadenosin/deoxycytidine kinase dgk BSU00150 deoxyguanosine kinase yaaH BSU00160 general stress protein, survival of ethanol stress, SafA-dependent spore coat yaaI BSU00170 general stress protein, similar to isochorismatase yaaJ BSU00180 tRNA specific adenosine