©Ferrata Storti Foundation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Genetical Society of Great Britain
Heredity 59 (1987) 151—160 The Genetical Society of Great Britain THEGENETICAL SOCIETY (Abstracts of papers presented at the TVVO HUNDRED AND FIFTH MEETING of the Society held on Friday, 14th and Saturday, 15th November 1986 at UNIVERSITY COLLEGE, LONDON) 1. Selection of somatic cell D. J. Porteous, P. A. Boyd, N. D. Hastie and hybrids with specific chromosome V. van Heyningen content for mapping the WAGR MAC Clinical and Population Cytogenetics Unit, Western General Hospital, Crewe Road, syndrome Edinburgh EH4 2XU. J. M. Fletcher, H. Morrison, J. A. Fantes, Clonedprobes for a number of available chromo- A. Seawright, S. Christie, D. J. Porteous, some ii assigned genes were used to define the N. D. Hastie and V. van Heyningen extent of deletions associated with the Wilms' MAC Clinical and Population Cytogenetics Unit, tumour, aniridia, genitourinary abnormalities and Western General Hospital, Crewe Road, mental retardation (WAGR) syndrome. Establish- Edinburgh EH4 2XU. ing reliable dosage studies for a number of different probes has proved difficult. We have therefore WAGR(Wilms tumour, aniridia, genitourinary abnormalities and mental retardation) syndrome concentrated on segregating the deleted chromo- is frequently associated with deletions on the short some 11 from a number of patients in somatic cell arm of chromosome 11. The deletions vary in size hybrids and analysing DNA from these to produce but always include part of band lipl3. To home a consistent map of chromosome lip. At the same in on the Wilms tumour and aniridia loci the end time we have determined the deletion breakpoints points of the different deletion breakpoints need at a molecular level and shown that the results are to be defined at the DNA level. -

Minireview Cilia and Flagella Revealed: from Flagellar Assembly
CORE Metadata, citation and similar papers at core.ac.uk Provided by Elsevier - Publisher Connector Cell, Vol. 117, 693–697, June 11, 2004, Copyright 2004 by Cell Press Cilia and Flagella Revealed: From Minireview Flagellar Assembly in Chlamydomonas to Human Obesity Disorders William J. Snell,* Junmin Pan, and Qian Wang This flow of materials is driven by the IFT machinery. Department of Cell Biology Flagellar proteins synthesized in the cell body are car- University of Texas Southwestern Medical School ried to the tip of the flagellum (the site of assembly of 5323 Harry Hines Boulevard the axoneme) by IFT particles, which are composed of Dallas, Texas 75390 at least 17 highly conserved proteins that form A and B complexes. The plus end-directed microtubule motor protein kinesin II is essential for movement of particles The recent identification in Chlamydomonas of the in- and their cargo toward the tip (anterograde transport) traflagellar transport machinery that assembles cilia of the flagellum, and a cytoplasmic dynein carries IFT and flagella has triggered a renaissance of interest in particles back to the cell body (retrograde transport). these organelles that transcends studies on their well- Thus, IFT particles function as constantly moving molec- characterized ability to move. New studies on several ular trucks on a closed loop. The tracks they travel on fronts have revealed that the machinery for flagellar are the microtubule doublets of the ciliary/flagellar axo- assembly/disassembly is regulated by homologs of neme, microtubule motors power them, and the individ- mitotic proteins, that cilia play essential roles in sensory ual structural components (e.g., microtubule subunits, transduction, and that mutations in cilia/basal body pro- dynein arms, and radial spoke proteins) of the cilium/ teins are responsible for cilia-related human disorders flagellum are their cargo. -

Dietary Supplementation with L-Arginine, Single Nucleotide
The Journal of Nutrition Commentary Dietary Supplementation with L-Arginine, Single Nucleotide Polymorphisms of Arginase Downloaded from https://academic.oup.com/jn/advance-article/doi/10.1093/jn/nxaa431/6131937 by University of Memphis - Library user on 10 February 2021 1 and 2, and Plasma L-Arginine Keith R Martin Center for Nutraceutical and Dietary Supplement Research, College of Health Sciences, University of Memphis, Memphis, TN, USA There are ∼3 million single nucleotide polymorphisms (SNPs) Arginine-dependent NO impacts the immune, neuronal, and between 2 unrelated individuals, with each SNP representing cardiovascular systems and is a potent vasodilator critical for a single modification in the genomic code and cumulatively vascular tone, blood pressure modulation, vascular permeabil- representing 0.1% human genetic diversity. Although many of ity, and angiogenesis (7–9). SNPs in the ARG1 gene are clinically these differences are silent, relatively recent data suggest that associated with cardiovascular diseases such as hypertension, around half of the various responses to dietary agents could cardiomyopathy, myocardial infarction, and carotid intima be related to genetic variation, a field coined as nutrigenetics. media thickness, and upregulation of arginase is implicated in These variations are of considerable interest in improving the vascular dysfunction (10, 11). Thus, dietary supplementation health of individuals and collectively mitigating the risk of with l-arginine could be warranted. chronic disease. In this issue of the Journal, Hannemann et The study by Hannemann et al. is the first to describe al. (1) describe a possible functional relation between arginase an association between genotypes of ARG1 and ARG2 with isoforms, important in the urea cycle and nitric oxide (NO) consequent circulating plasma concentrations of l-arginine. -

Mesenchymal Stem Cells Prevent the Progression of Diabetic
Lee et al. Experimental & Molecular Medicine (2019) 51:77 https://doi.org/10.1038/s12276-019-0268-5 Experimental & Molecular Medicine ARTICLE Open Access Mesenchymal stem cells prevent the progression of diabetic nephropathy by improving mitochondrial function in tubular epithelial cells Seung Eun Lee1,2,7,JungEunJang1,2,8,HyunSikKim2,MinKyoJung2,MyoungSeokKo2,Mi-OkKim2, Hye Sun Park2, Wonil Oh3, Soo Jin Choi3,HyeJinJin3, Sang-Yeob Kim4,YunJaeKim2,SeongWhoKim5, Min Kyung Kim5, Chang Ohk Sung6, Chan-Gi Pack 2,Ki-UpLee1,2 and Eun Hee Koh1,2 Abstract The administration of mesenchymal stem cells (MSCs) was shown to attenuate overt as well as early diabetic nephropathy in rodents, but the underlying mechanism of this beneficial effect is largely unknown. Inflammation and mitochondrial dysfunction are major pathogenic factors in diabetic nephropathy. In this study, we found that the repeated administration of MSCs prevents albuminuria and injury to tubular epithelial cells (TECs), an important element in the progression of diabetic nephropathy, by improving mitochondrial function. The expression of M1 macrophage markers was significantly increased in diabetic kidneys compared with that in control kidneys. Interestingly, the expression of arginase-1 (Arg1), an important M2 macrophage marker, was reduced in diabetic kidneys and increased by MSC treatment. In cultured TECs, conditioned media from lipopolysaccharide-activated 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; macrophages reduced peroxisomal proliferator-activated receptor gamma coactivator 1α (Pgc1a) expression and impaired mitochondrial function. The coculture of macrophages with MSCs increased and decreased the expression of Arg1 and M1 markers, respectively. Treatment with conditioned media from cocultured macrophages prevented activated macrophage-induced mitochondrial dysfunction in TECs. -

Anti-Cdk8 Antibody Produced in Rabbit (C0238)
ANTI-CDK8 Developed in Rabbit, Affinity Isolated Antibody Product Number C 0238 Product Description Reagent Anti-Cyclin-Dependent Kinase 8 (CDK8) is developed in Anti-CDK8, at approximately 1 mg/ml, is supplied as a rabbit using a synthetic peptide corresponding to the solution in phosphate buffered saline, pH 7.4 containing C-terminal region (aa 451-464) of human CDK8. The 0.2% BSA and 15 mM sodium azide. antibody is purified by protein A affinity chromatography. Anti-CDK8 specifically recognizes a 54 kDa protein Precautions and Disclaimer identified as cyclin-dependent kinase 8 (CDK8). Anti- Due to the sodium azide content, a material safety data CDK8 does not crossreact with the other members of sheet (MSDS) for this product has been sent to the CDK family. It detects human, mouse and rat CDK8. It attention of the safety officer of your institution. Consult is used in immunoblotting, immunoprecipitation and the MSDS for information regarding hazardous and safe immunofluorescence applications. handling practices. Cyclin-dependent kinase 8 (CDK8) is a 464-amino acid Storage/Stability protein containing the sequence motifs and 11 Store at –20 °C. For extended storage, upon initial subdomains characteristic of a serine/threonine-specific thawing, freeze the solution in working aliquots. kinase.1 CDKs become activated through binding to Avoid repeated freezing and thawing to prevent cyclins, formation of cyclin-CDK complexes and denaturing the antibody. Storage in “frost-free” reversible phosphorylation reactions. Cyclin-CDK freezers is also not recommended. If slight complexes directly control progression through G1, S, turbidity occurs upon prolonged storage, clarify the G2 and M phases of the cell division cycle. -

Chuanxiong Rhizoma Compound on HIF-VEGF Pathway and Cerebral Ischemia-Reperfusion Injury’S Biological Network Based on Systematic Pharmacology
ORIGINAL RESEARCH published: 25 June 2021 doi: 10.3389/fphar.2021.601846 Exploring the Regulatory Mechanism of Hedysarum Multijugum Maxim.-Chuanxiong Rhizoma Compound on HIF-VEGF Pathway and Cerebral Ischemia-Reperfusion Injury’s Biological Network Based on Systematic Pharmacology Kailin Yang 1†, Liuting Zeng 1†, Anqi Ge 2†, Yi Chen 1†, Shanshan Wang 1†, Xiaofei Zhu 1,3† and Jinwen Ge 1,4* Edited by: 1 Takashi Sato, Key Laboratory of Hunan Province for Integrated Traditional Chinese and Western Medicine on Prevention and Treatment of 2 Tokyo University of Pharmacy and Life Cardio-Cerebral Diseases, Hunan University of Chinese Medicine, Changsha, China, Galactophore Department, The First 3 Sciences, Japan Hospital of Hunan University of Chinese Medicine, Changsha, China, School of Graduate, Central South University, Changsha, China, 4Shaoyang University, Shaoyang, China Reviewed by: Hui Zhao, Capital Medical University, China Background: Clinical research found that Hedysarum Multijugum Maxim.-Chuanxiong Maria Luisa Del Moral, fi University of Jaén, Spain Rhizoma Compound (HCC) has de nite curative effect on cerebral ischemic diseases, *Correspondence: such as ischemic stroke and cerebral ischemia-reperfusion injury (CIR). However, its Jinwen Ge mechanism for treating cerebral ischemia is still not fully explained. [email protected] †These authors share first authorship Methods: The traditional Chinese medicine related database were utilized to obtain the components of HCC. The Pharmmapper were used to predict HCC’s potential targets. Specialty section: The CIR genes were obtained from Genecards and OMIM and the protein-protein This article was submitted to interaction (PPI) data of HCC’s targets and IS genes were obtained from String Ethnopharmacology, a section of the journal database. -

Investigation of COVID-19 Comorbidities Reveals Genes and Pathways Coincident with the SARS-Cov-2 Viral Disease
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.21.306720; this version posted September 21, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Title: Investigation of COVID-19 comorbidities reveals genes and pathways coincident with the SARS-CoV-2 viral disease. Authors: Mary E. Dolan1*,2, David P. Hill1,2, Gaurab Mukherjee2, Monica S. McAndrews2, Elissa J. Chesler2, Judith A. Blake2 1 These authors contributed equally and should be considered co-first authors * Corresponding author [email protected] 2 The Jackson Laboratory, 600 Main St, Bar Harbor, ME 04609, USA Abstract: The emergence of the SARS-CoV-2 virus and subsequent COVID-19 pandemic initiated intense research into the mechanisms of action for this virus. It was quickly noted that COVID-19 presents more seriously in conjunction with other human disease conditions such as hypertension, diabetes, and lung diseases. We conducted a bioinformatics analysis of COVID-19 comorbidity-associated gene sets, identifying genes and pathways shared among the comorbidities, and evaluated current knowledge about these genes and pathways as related to current information about SARS-CoV-2 infection. We performed our analysis using GeneWeaver (GW), Reactome, and several biomedical ontologies to represent and compare common COVID- 19 comorbidities. Phenotypic analysis of shared genes revealed significant enrichment for immune system phenotypes and for cardiovascular-related phenotypes, which might point to alleles and phenotypes in mouse models that could be evaluated for clues to COVID-19 severity. -

MECHANISMS in ENDOCRINOLOGY: Novel Genetic Causes of Short Stature
J M Wit and others Genetics of short stature 174:4 R145–R173 Review MECHANISMS IN ENDOCRINOLOGY Novel genetic causes of short stature 1 1 2 2 Jan M Wit , Wilma Oostdijk , Monique Losekoot , Hermine A van Duyvenvoorde , Correspondence Claudia A L Ruivenkamp2 and Sarina G Kant2 should be addressed to J M Wit Departments of 1Paediatrics and 2Clinical Genetics, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, Email The Netherlands [email protected] Abstract The fast technological development, particularly single nucleotide polymorphism array, array-comparative genomic hybridization, and whole exome sequencing, has led to the discovery of many novel genetic causes of growth failure. In this review we discuss a selection of these, according to a diagnostic classification centred on the epiphyseal growth plate. We successively discuss disorders in hormone signalling, paracrine factors, matrix molecules, intracellular pathways, and fundamental cellular processes, followed by chromosomal aberrations including copy number variants (CNVs) and imprinting disorders associated with short stature. Many novel causes of GH deficiency (GHD) as part of combined pituitary hormone deficiency have been uncovered. The most frequent genetic causes of isolated GHD are GH1 and GHRHR defects, but several novel causes have recently been found, such as GHSR, RNPC3, and IFT172 mutations. Besides well-defined causes of GH insensitivity (GHR, STAT5B, IGFALS, IGF1 defects), disorders of NFkB signalling, STAT3 and IGF2 have recently been discovered. Heterozygous IGF1R defects are a relatively frequent cause of prenatal and postnatal growth retardation. TRHA mutations cause a syndromic form of short stature with elevated T3/T4 ratio. Disorders of signalling of various paracrine factors (FGFs, BMPs, WNTs, PTHrP/IHH, and CNP/NPR2) or genetic defects affecting cartilage extracellular matrix usually cause disproportionate short stature. -

Chromosome 6 Abnormalities in Ovarian Surface Epithelial Tumors of Borderline Malignancy Suggest a Genetic Continuum in the Progression Model of Ovarian Neoplasms1
3404 Vol. 7, 3404–3409, November 2001 Clinical Cancer Research Chromosome 6 Abnormalities in Ovarian Surface Epithelial Tumors of Borderline Malignancy Suggest a Genetic Continuum in the Progression Model of Ovarian Neoplasms1 Maria Grazia Tibiletti, Barbara Bernasconi, Conclusion: Alterations in the above-mentioned inter- Daniela Furlan, Paola Bressan, Roberta Cerutti, val are a common finding in advanced ovarian carcinomas but also in benign ovarian cysts, implying that some tumors Carla Facco, Massimo Franchi, Cristina Riva, of borderline malignancy may arise from benign tumors and Raffaella Cinquetti, Carlo Capella, and that malignant ones may evolve from tumors of borderline Roberto Taramelli2 malignancy. Genes located in 6q27 seem to be crucial for this Laboratorio di Anatomia Patologica Ospedale di Circolo, Fondazione mechanism of early events in ovarian tumorigenesis. Macchi, 21100 Varese [M. G. T., C. F., C. R.]; and Dipartimento di Scienze Cliniche e Biologiche [B. B., D. F., P. B., R. Ce., C. C.], INTRODUCTION Clinica Ostetrica e Ginecologica [M. F.], and Dipartimento di Biologia Strutturale e Funzionale [R. Ci., R. T.], Universita` Ovarian cancer represents the most lethal of the gyneco- dell’Insubria, 21100 Varese, Italy logical tumors, and its pathogenesis is poorly understood. More than 80% of malignant tumors of the ovary are of epithelial origin and arise from the surface epithelium (1). Ovarian surface ABSTRACT epithelial tumors are divided into three histological subtypes: Purpose: We used conventional cytogenetics, molecular benign cystoadenoma, TBM,3 and AC, which are related to a cytogenetics, and molecular genetic analyses to study the different biological behavior (1). The molecular events under- pattern of allelic loss on chromosome 6q in a cohort of lying the development of ovarian tumorigenesis are still largely borderline epithelial ovarian tumors. -

Regulatory Functions of the Mediator Kinases CDK8 and CDK19 Charli B
TRANSCRIPTION 2019, VOL. 10, NO. 2, 76–90 https://doi.org/10.1080/21541264.2018.1556915 REVIEW Regulatory functions of the Mediator kinases CDK8 and CDK19 Charli B. Fant and Dylan J. Taatjes Department of Biochemistry, University of Colorado, Boulder, CO, USA ABSTRACT ARTICLE HISTORY The Mediator-associated kinases CDK8 and CDK19 function in the context of three additional Received 19 September 2018 proteins: CCNC and MED12, which activate CDK8/CDK19 kinase function, and MED13, which Revised 13 November 2018 enables their association with the Mediator complex. The Mediator kinases affect RNA polymerase Accepted 20 November 2018 II (pol II) transcription indirectly, through phosphorylation of transcription factors and by control- KEYWORDS ling Mediator structure and function. In this review, we discuss cellular roles of the Mediator Mediator kinase; enhancer; kinases and mechanisms that enable their biological functions. We focus on sequence-specific, transcription; RNA DNA-binding transcription factors and other Mediator kinase substrates, and how CDK8 or CDK19 polymerase II; chromatin may enable metabolic and transcriptional reprogramming through enhancers and chromatin looping. We also summarize Mediator kinase inhibitors and their therapeutic potential. Throughout, we note conserved and divergent functions between yeast and mammalian CDK8, and highlight many aspects of kinase module function that remain enigmatic, ranging from potential roles in pol II promoter-proximal pausing to liquid-liquid phase separation. Introduction and MED13L associate in a mutually exclusive fash- ion with MED12 and MED13 [12], and their poten- The CDK8 kinase exists in a 600 kDa complex tial functional distinctions remain unclear. known as the CDK8 module, which consists of four CDK8 is considered both an oncogene [13–15] proteins (CDK8, CCNC, MED12, MED13). -
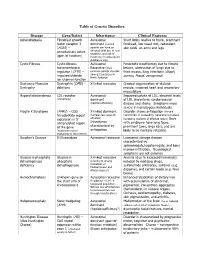
Table of Genetic Disorders Disease Gene/Defect Inheritance Clinical
Table of Genetic Disorders Disease Gene/Defect Inheritance Clinical Features Achondroplasia Fibroblast growth Autosomal Short limbs relative to trunk, prominent factor receptor 3 dominant (normal forehead, low nasal root, redundant (FGR3) – parents can have an skin folds on arms and legs constitutively active affected child due to new mutation, and risk of (gain of function) recurrence in subsequent children is low) Cystic Fibrosis Cystic fibrosis Autosomal Pancreatic insufficiency due to fibrotic transmembrane Recessive (most lesions, obstruction of lungs due to regulator (CFTR) – common genetic disorder thick mucus, lung infections (Staph, impaired chloride among Caucasians in aureus, Pseud. aeruginosa) North America) ion channel function Duchenne Muscular Dystrophin (DMD) - X-linked recessive Gradual degeneration of skeletal Dystrophy deletions muscle, impaired heart and respiratory musculature Hypercholesterolemia LDL receptor Autosomal Impaired uptake of LDL, elevated levels (commonly) dominant of LDL cholesterol, cardiovascular (haploinsufficiency) disease and stroke. Symptoms more severe in homozygous individuals Fragile X Syndrome (FMR1) – CGG X-linked dominant Disorder shows anticipation (female trinucleotide repeat (females less severely transmitters in succeeding generations produce expansion in 5’ affected) increasing numbers of affected males) Boys untranslated region Inheritance with syndrome have long faces, of the gene characterized by prominent jaws, large ears, and are (expansion occurs anticipation likely to be mentally retarded. exclusively in the mother) Gaucher’s Disease Β-Glucosidase Autosomal recessive Lysosomal storage disease characterized by splenomegaly,hepatomegaly, and bone marrow infiltration. Neurological symptoms are not common Glucose 6-phosphate Glucose 6- X-linked recessive Anemia (due to increased hemolysis) dehydrogenase phosphate (prominent among induced by oxidizing drugs, deficiency dehydrogenase individuals of sulfonamide antibiotics, sulfones (e.g. -

The Retinitis Pigmentosa 1 Protein Is a Photoreceptor Microtubule-Associated Protein
The Journal of Neuroscience, July 21, 2004 • 24(29):6427–6436 • 6427 Neurobiology of Disease The Retinitis Pigmentosa 1 Protein Is a Photoreceptor Microtubule-Associated Protein Qin Liu,1 Jian Zuo,2 and Eric A. Pierce1 1F. M. Kirby Center for Molecular Ophthalmology, Scheie Eye Institute, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania 19104, and 2Department of Developmental Neurobiology, St. Jude Children’s Research Hospital, Memphis, Tennessee 38105 The outer segments of rod and cone photoreceptor cells are highly specialized sensory cilia made up of hundreds of membrane discs stacked into an orderly array along the photoreceptor axoneme. It is not known how the alignment of the outer segment discs is controlled, although it has been suggested that the axoneme may play a role in this process. Mutations in the retinitis pigmentosa 1 (RP1) gene are a common cause of retinitis pigmentosa (RP). Disruption of the Rp1 gene in mice causes misorientation of outer segment discs, suggesting a role for RP1 in outer segment organization. Here, we show that the RP1 protein is part of the photoreceptor axoneme. Amino acids 28–228 of RP1, which share limited homology with the microtubule-binding domains of the neuronal microtubule-associated protein (MAP) doublecortin, mediate the interaction between RP1 and microtubules, indicating that the putative doublecortin (DCX) domains in RP1 are functional. The N-terminal portion of RP1 stimulates the formation of microtubules in vitro and stabilizes cytoplas- mic microtubules in heterologous cells. Evaluation of photoreceptor axonemes from mice with targeted disruptions of the Rp1 gene shows that Rp1 proteins that contain the DCX domains also help control axoneme length and stability in vivo.