Gain of Chromosome 6P Is an Infrequent Cause of Increased PIM1 Expression in B-Cell Non-Hodgkin’S Lymphomas
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Genetical Society of Great Britain
Heredity 59 (1987) 151—160 The Genetical Society of Great Britain THEGENETICAL SOCIETY (Abstracts of papers presented at the TVVO HUNDRED AND FIFTH MEETING of the Society held on Friday, 14th and Saturday, 15th November 1986 at UNIVERSITY COLLEGE, LONDON) 1. Selection of somatic cell D. J. Porteous, P. A. Boyd, N. D. Hastie and hybrids with specific chromosome V. van Heyningen content for mapping the WAGR MAC Clinical and Population Cytogenetics Unit, Western General Hospital, Crewe Road, syndrome Edinburgh EH4 2XU. J. M. Fletcher, H. Morrison, J. A. Fantes, Clonedprobes for a number of available chromo- A. Seawright, S. Christie, D. J. Porteous, some ii assigned genes were used to define the N. D. Hastie and V. van Heyningen extent of deletions associated with the Wilms' MAC Clinical and Population Cytogenetics Unit, tumour, aniridia, genitourinary abnormalities and Western General Hospital, Crewe Road, mental retardation (WAGR) syndrome. Establish- Edinburgh EH4 2XU. ing reliable dosage studies for a number of different probes has proved difficult. We have therefore WAGR(Wilms tumour, aniridia, genitourinary abnormalities and mental retardation) syndrome concentrated on segregating the deleted chromo- is frequently associated with deletions on the short some 11 from a number of patients in somatic cell arm of chromosome 11. The deletions vary in size hybrids and analysing DNA from these to produce but always include part of band lipl3. To home a consistent map of chromosome lip. At the same in on the Wilms tumour and aniridia loci the end time we have determined the deletion breakpoints points of the different deletion breakpoints need at a molecular level and shown that the results are to be defined at the DNA level. -

MECHANISMS in ENDOCRINOLOGY: Novel Genetic Causes of Short Stature
J M Wit and others Genetics of short stature 174:4 R145–R173 Review MECHANISMS IN ENDOCRINOLOGY Novel genetic causes of short stature 1 1 2 2 Jan M Wit , Wilma Oostdijk , Monique Losekoot , Hermine A van Duyvenvoorde , Correspondence Claudia A L Ruivenkamp2 and Sarina G Kant2 should be addressed to J M Wit Departments of 1Paediatrics and 2Clinical Genetics, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, Email The Netherlands [email protected] Abstract The fast technological development, particularly single nucleotide polymorphism array, array-comparative genomic hybridization, and whole exome sequencing, has led to the discovery of many novel genetic causes of growth failure. In this review we discuss a selection of these, according to a diagnostic classification centred on the epiphyseal growth plate. We successively discuss disorders in hormone signalling, paracrine factors, matrix molecules, intracellular pathways, and fundamental cellular processes, followed by chromosomal aberrations including copy number variants (CNVs) and imprinting disorders associated with short stature. Many novel causes of GH deficiency (GHD) as part of combined pituitary hormone deficiency have been uncovered. The most frequent genetic causes of isolated GHD are GH1 and GHRHR defects, but several novel causes have recently been found, such as GHSR, RNPC3, and IFT172 mutations. Besides well-defined causes of GH insensitivity (GHR, STAT5B, IGFALS, IGF1 defects), disorders of NFkB signalling, STAT3 and IGF2 have recently been discovered. Heterozygous IGF1R defects are a relatively frequent cause of prenatal and postnatal growth retardation. TRHA mutations cause a syndromic form of short stature with elevated T3/T4 ratio. Disorders of signalling of various paracrine factors (FGFs, BMPs, WNTs, PTHrP/IHH, and CNP/NPR2) or genetic defects affecting cartilage extracellular matrix usually cause disproportionate short stature. -

Chromosome 6 Abnormalities in Ovarian Surface Epithelial Tumors of Borderline Malignancy Suggest a Genetic Continuum in the Progression Model of Ovarian Neoplasms1
3404 Vol. 7, 3404–3409, November 2001 Clinical Cancer Research Chromosome 6 Abnormalities in Ovarian Surface Epithelial Tumors of Borderline Malignancy Suggest a Genetic Continuum in the Progression Model of Ovarian Neoplasms1 Maria Grazia Tibiletti, Barbara Bernasconi, Conclusion: Alterations in the above-mentioned inter- Daniela Furlan, Paola Bressan, Roberta Cerutti, val are a common finding in advanced ovarian carcinomas but also in benign ovarian cysts, implying that some tumors Carla Facco, Massimo Franchi, Cristina Riva, of borderline malignancy may arise from benign tumors and Raffaella Cinquetti, Carlo Capella, and that malignant ones may evolve from tumors of borderline Roberto Taramelli2 malignancy. Genes located in 6q27 seem to be crucial for this Laboratorio di Anatomia Patologica Ospedale di Circolo, Fondazione mechanism of early events in ovarian tumorigenesis. Macchi, 21100 Varese [M. G. T., C. F., C. R.]; and Dipartimento di Scienze Cliniche e Biologiche [B. B., D. F., P. B., R. Ce., C. C.], INTRODUCTION Clinica Ostetrica e Ginecologica [M. F.], and Dipartimento di Biologia Strutturale e Funzionale [R. Ci., R. T.], Universita` Ovarian cancer represents the most lethal of the gyneco- dell’Insubria, 21100 Varese, Italy logical tumors, and its pathogenesis is poorly understood. More than 80% of malignant tumors of the ovary are of epithelial origin and arise from the surface epithelium (1). Ovarian surface ABSTRACT epithelial tumors are divided into three histological subtypes: Purpose: We used conventional cytogenetics, molecular benign cystoadenoma, TBM,3 and AC, which are related to a cytogenetics, and molecular genetic analyses to study the different biological behavior (1). The molecular events under- pattern of allelic loss on chromosome 6q in a cohort of lying the development of ovarian tumorigenesis are still largely borderline epithelial ovarian tumors. -
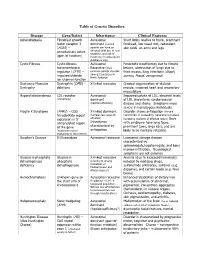
Table of Genetic Disorders Disease Gene/Defect Inheritance Clinical
Table of Genetic Disorders Disease Gene/Defect Inheritance Clinical Features Achondroplasia Fibroblast growth Autosomal Short limbs relative to trunk, prominent factor receptor 3 dominant (normal forehead, low nasal root, redundant (FGR3) – parents can have an skin folds on arms and legs constitutively active affected child due to new mutation, and risk of (gain of function) recurrence in subsequent children is low) Cystic Fibrosis Cystic fibrosis Autosomal Pancreatic insufficiency due to fibrotic transmembrane Recessive (most lesions, obstruction of lungs due to regulator (CFTR) – common genetic disorder thick mucus, lung infections (Staph, impaired chloride among Caucasians in aureus, Pseud. aeruginosa) North America) ion channel function Duchenne Muscular Dystrophin (DMD) - X-linked recessive Gradual degeneration of skeletal Dystrophy deletions muscle, impaired heart and respiratory musculature Hypercholesterolemia LDL receptor Autosomal Impaired uptake of LDL, elevated levels (commonly) dominant of LDL cholesterol, cardiovascular (haploinsufficiency) disease and stroke. Symptoms more severe in homozygous individuals Fragile X Syndrome (FMR1) – CGG X-linked dominant Disorder shows anticipation (female trinucleotide repeat (females less severely transmitters in succeeding generations produce expansion in 5’ affected) increasing numbers of affected males) Boys untranslated region Inheritance with syndrome have long faces, of the gene characterized by prominent jaws, large ears, and are (expansion occurs anticipation likely to be mentally retarded. exclusively in the mother) Gaucher’s Disease Β-Glucosidase Autosomal recessive Lysosomal storage disease characterized by splenomegaly,hepatomegaly, and bone marrow infiltration. Neurological symptoms are not common Glucose 6-phosphate Glucose 6- X-linked recessive Anemia (due to increased hemolysis) dehydrogenase phosphate (prominent among induced by oxidizing drugs, deficiency dehydrogenase individuals of sulfonamide antibiotics, sulfones (e.g. -

Molecular Cytogenetic Characterisation of a Novel De Novo Ring
Pace et al. Molecular Cytogenetics (2017) 10:9 DOI 10.1186/s13039-017-0311-y CASEREPORT Open Access Molecular cytogenetic characterisation of a novel de novo ring chromosome 6 involving a terminal 6p deletion and terminal 6q duplication in the different arms of the same chromosome Nikolai Paul Pace1, Frideriki Maggouta2, Melissa Twigden2 and Isabella Borg1,3,4* Abstract Background: Ring chromosome 6 is a rare sporadic chromosomal abnormality, associated with extreme variability in clinical phenotypes. Most ring chromosomes are known to have deletions on one or both chromosomal arms. Here, we report an atypical and unique ring chromosome 6 involving both a distal deletion and a distal duplication on the different arms of the same chromosome. Case presentation: In a patient with intellectual disability, short stature, microcephaly, facial dysmorphology, congenital heart defects and renovascular disease, a ring chromosome 6 was characterised using array-CGH and dual-colour FISH. The de-novo ring chromosome 6 involved a 1.8 Mb terminal deletion in the distal short arm and a 2.5 Mb duplication in the distal long arm of the same chromosome 6. This results in monosomy for the region 6pter to 6p25.3 and trisomy for the region 6q27 to 6qter. Analysis of genes in these chromosomal regions suggests that haploinsufficiency for FOXC1 and GMDS genes accounts for the cardiac and neurodevelopmental phenotypes in the proband. The ring chromosome 6 reported here is atypical as it involves a unique duplication of the distal long arm. Furthermore, the presence of renovascular disease is also a unique feature identified in this patient. -

WNT16 Is a New Marker of Senescence
Table S1. A. Complete list of 177 genes overexpressed in replicative senescence Value Gene Description UniGene RefSeq 2.440 WNT16 wingless-type MMTV integration site family, member 16 (WNT16), transcript variant 2, mRNA. Hs.272375 NM_016087 2.355 MMP10 matrix metallopeptidase 10 (stromelysin 2) (MMP10), mRNA. Hs.2258 NM_002425 2.344 MMP3 matrix metallopeptidase 3 (stromelysin 1, progelatinase) (MMP3), mRNA. Hs.375129 NM_002422 2.300 HIST1H2AC Histone cluster 1, H2ac Hs.484950 2.134 CLDN1 claudin 1 (CLDN1), mRNA. Hs.439060 NM_021101 2.119 TSPAN13 tetraspanin 13 (TSPAN13), mRNA. Hs.364544 NM_014399 2.112 HIST2H2BE histone cluster 2, H2be (HIST2H2BE), mRNA. Hs.2178 NM_003528 2.070 HIST2H2BE histone cluster 2, H2be (HIST2H2BE), mRNA. Hs.2178 NM_003528 2.026 DCBLD2 discoidin, CUB and LCCL domain containing 2 (DCBLD2), mRNA. Hs.203691 NM_080927 2.007 SERPINB2 serpin peptidase inhibitor, clade B (ovalbumin), member 2 (SERPINB2), mRNA. Hs.594481 NM_002575 2.004 HIST2H2BE histone cluster 2, H2be (HIST2H2BE), mRNA. Hs.2178 NM_003528 1.989 OBFC2A Oligonucleotide/oligosaccharide-binding fold containing 2A Hs.591610 1.962 HIST2H2BE histone cluster 2, H2be (HIST2H2BE), mRNA. Hs.2178 NM_003528 1.947 PLCB4 phospholipase C, beta 4 (PLCB4), transcript variant 2, mRNA. Hs.472101 NM_182797 1.934 PLCB4 phospholipase C, beta 4 (PLCB4), transcript variant 1, mRNA. Hs.472101 NM_000933 1.933 KRTAP1-5 keratin associated protein 1-5 (KRTAP1-5), mRNA. Hs.534499 NM_031957 1.894 HIST2H2BE histone cluster 2, H2be (HIST2H2BE), mRNA. Hs.2178 NM_003528 1.884 CYTL1 cytokine-like 1 (CYTL1), mRNA. Hs.13872 NM_018659 tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain (TNFRSF10D), 1.848 TNFRSF10D Hs.213467 NM_003840 mRNA. -

Isolation of Chromosome-Specific Ests by Microdissection-Mediated Cdna Capture Edgardo Gracia, 1-3 Michael E
Downloaded from genome.cshlp.org on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press RESEARCH Isolation of Chromosome-Specific ESTs by Microdissection-Mediated cDNA Capture Edgardo Gracia, 1-3 Michael E. Ray, 1-3 Mihael H. Polymeropoulos, 4 Anindya Dehejia, 4 Paul S. Meltzer, 3 and Jeffrey M. Trent 3's 1Department of Human Genetics, The University of Michigan Medical School, Medical Science II M4708, Ann Arbor, Michigan 48109; 3Laboratory of Cancer Genetics, 4 Laboratory of Genetic Disease Research, National Center for Human Genome Research, National Institutes of Health, Bethesda, Maryland 20892 Despite dramatic advances in the identification of human expressed sequence tags (ESTs), techniques that facilitate isolation of chromosome or chromosome band-specific ESTs would be of considerable value. This report demonstrates the feasibility of identifying chromosome-specific ESTs following microdissection of a single-copy chromosome region. For this study, a reduced complexity cDNA library was linkered and hybridized to normal human metaphase chromosomes. After stringency washes, the entire long arm of chromosome 6 (6q) was microdissected. Following PCR amplification using linker-specific primers, captured cDNAs were subcloned and 187 individual clones picked at random. These 187 clones were then sorted by filter cross-hybridization into 34 unique groups. Of these 34 groups, 19 (56%) mapped to chromosome 6 by Southern blot. We identified three previously known genes, human cytovillin (ezrin) mapped previously to 6q25-26, human cardiac gap junction protein (connexin 43) mapped previously to 6q21-23.2 and prolyloligopeptidase, which had not been mapped previously. BLASTN identified three clone groups with homology to known ESTs and 12 representing novel cDNA sequences. -

Of Small Intestine Harboring Driver Gene Mutations: a Case Report and a Literature Review
1161 Case Report A rare multiple primary sarcomatoid carcinoma (SCA) of small intestine harboring driver gene mutations: a case report and a literature review Zhu Zhu1#, Xinyi Liu2#, Wenliang Li1, Zhengqi Wen1, Xiang Ji1, Ruize Zhou1, Xiaoyu Tuo3, Yaru Chen2, Xian Gong2, Guifeng Liu2, Yanqing Zhou2, Shifu Chen2, Lele Song2#^, Jian Huang1 1Department of Oncology, First Affiliated Hospital of Kunming Medical University, Kunming, China; 2HaploX Biotechnology, Shenzhen, China; 3Department of Pathology, First Affiliated Hospital of Kunming Medical University, Kunming, China #These authors contributed equally to this work. Correspondence to: Jian Huang. Department of Oncology, First Affiliated Hospital of Kunming Medical University, No. 295, Xichang Road, Kunming 560032, Yunnan Province, China. Email: [email protected]; Lele Song. HaploX Biotechnology, 8th floor, Auto Electric Power Building, Songpingshan Road, Nanshan District, Shenzhen 518057, Guangdong Province, China. Email: [email protected]. Abstract: Primary sarcomatoid carcinoma (SCA) is a type of rare tumor consisting of both malignant epithelial and mesenchymal components. Only 32 cases of SCA of the small bowel have been reported in the literature to date. Due to its rarity and complexity, this cancer has not been genetically studied and its diagnosis and treatment remain difficult. Here we report a 54-year-old male underwent emergency surgical resection in the small intestine due to severe obstruction and was diagnosed with multiple SCA based on postoperative pathological examination. Over 100 polypoid tumors scattered along his whole jejunum and proximal ileum. Chemotherapy (IFO+Epirubicin) was performed after surgery while the patient died two months after the surgery due to severe malnutrition. Whole-exome sequencing was performed for the tumor tissue with normal tissue as the control. -

The Kinesin Spindle Protein Inhibitor Filanesib Enhances the Activity of Pomalidomide and Dexamethasone in Multiple Myeloma
Plasma Cell Disorders SUPPLEMENTARY APPENDIX The kinesin spindle protein inhibitor filanesib enhances the activity of pomalidomide and dexamethasone in multiple myeloma Susana Hernández-García, 1 Laura San-Segundo, 1 Lorena González-Méndez, 1 Luis A. Corchete, 1 Irena Misiewicz- Krzeminska, 1,2 Montserrat Martín-Sánchez, 1 Ana-Alicia López-Iglesias, 1 Esperanza Macarena Algarín, 1 Pedro Mogollón, 1 Andrea Díaz-Tejedor, 1 Teresa Paíno, 1 Brian Tunquist, 3 María-Victoria Mateos, 1 Norma C Gutiérrez, 1 Elena Díaz- Rodriguez, 1 Mercedes Garayoa 1* and Enrique M Ocio 1* 1Centro Investigación del Cáncer-IBMCC (CSIC-USAL) and Hospital Universitario-IBSAL, Salamanca, Spain; 2National Medicines Insti - tute, Warsaw, Poland and 3Array BioPharma, Boulder, Colorado, USA *MG and EMO contributed equally to this work ©2017 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol. 2017.168666 Received: March 13, 2017. Accepted: August 29, 2017. Pre-published: August 31, 2017. Correspondence: [email protected] MATERIAL AND METHODS Reagents and drugs. Filanesib (F) was provided by Array BioPharma Inc. (Boulder, CO, USA). Thalidomide (T), lenalidomide (L) and pomalidomide (P) were purchased from Selleckchem (Houston, TX, USA), dexamethasone (D) from Sigma-Aldrich (St Louis, MO, USA) and bortezomib from LC Laboratories (Woburn, MA, USA). Generic chemicals were acquired from Sigma Chemical Co., Roche Biochemicals (Mannheim, Germany), Merck & Co., Inc. (Darmstadt, Germany). MM cell lines, patient samples and cultures. Origin, authentication and in vitro growth conditions of human MM cell lines have already been characterized (17, 18). The study of drug activity in the presence of IL-6, IGF-1 or in co-culture with primary bone marrow mesenchymal stromal cells (BMSCs) or the human mesenchymal stromal cell line (hMSC–TERT) was performed as described previously (19, 20). -

Orientation of Loci Within the Human Major Histocompatibility Complex by Chromosomal in Situ Hybridization (Human Leukocyte Antigen/Gene Mapping) CYNTHIA C
Proc. Nati. Acad. Sci. USA Vol. 81, pp. 2816-2820, May 1984 Genetics Orientation of loci within the human major histocompatibility complex by chromosomal in situ hybridization (human leukocyte antigen/gene mapping) CYNTHIA C. MORTON*t, ILAN R. KIRSCHt, WALTER E. NANCE*, GLEN A. EVANS§, ALAN J. KORMAN¶, AND JACK L. STROMINGER¶ *Department of Human Genetics, Medical College of Virginia, Virginia Commonwealth University, Richmond, VA 23298; tNational Cancer Institute, Navy Medical Oncology Branch, The National Naval Medical Center, Building 1, Room 415, Bethesda, MD 20814; §The Cancer Biology Laboratory, The Salk Institute, San Diego, CA 92138; and ¶Fairchild Biochemistry Building, Harvard University, 7 Divinity Avenue, Cambridge, MA 02138 Contributed by Jack L. Strominger, December 29, 1983 ABSTRACT We have determined the localization and ori- with closely linked markers such as glyoxylase 1 (GLO 1) entation of two genetic probes within the human major histo- have permitted inferences to be drawn about the order and compatibility complex by chromosomal in situ hybridization. map distances between marker loci on the proximal portion Our data indicate that a cloned genomic probe cross-hybridiz- of the short arm of chromosome 6, as follows: GLO 1-4 ing to HLA-A, -B, and -C heavy chain loci is homologous to centimorgans (cM)-HLA-DR-0.3 cM-BF, C4B, C4A, sequences located on chromosome 6 at band p21.3 while a sub- C2-0.7 cM-HLA-B-0.1 cM-HLA-C-0.7 cM-HLA-A clone of the genomic HLA-DR a-chain gene corresponding to (16-18). the nonpolymorphic p34 protein is homologous to sequences in Improvements in chromosomal in situ hybridization have band 6p21.1. -

The Role of PANE1 As a Model for Minor Histocompatibility Restricted Antigen in Hematopoietic Stem Cell Transplantation for Leukemic Patients
Hammad et al. SECI Oncology 2020 (2) DOI: The role of PANE1 as a model for minor histocompatibility restricted antigen in hematopoietic stem cell transplantation for leukemic patients Hammad MH 1, Elfishawi SM2, Zahran AM3, Abd El Fattah RM4, Bakry RM5 1 Assistant lecturer in Clinical Pathology Department, South Egypt Cancer Institute, Assiut University 2 Lecturer in Clinical Pathology Department, National Cancer Institute, Cairo University 3 Assistant professor in Clinical Pathology Department, South Egypt Cancer Institute, Assiut University 4 Consultant in Medical Oncology Department, National Cancer Institute, Cairo University 5 Professor in Clinical Pathology Department, South Egypt Cancer Institute, Assiut University e-mail: [email protected] Abstract Background: In hematopoietic stem cell transplantation (HSCT), minor histocompatibility antigens (MiHA) can be used as tools for immunotherapy especially if they are restricted to hematopoietic cells like PANE1 MiHA. Purpose of study: We aimed at studying PANE1 as MiHA model in human leucocyte antigen (HLA) matched related Egyptian HSCT patients regarding its frequency and the effect of mismatching in PANE1 on HSCT outcomes. Methods: Ninety-six patient/donor pairs were studied for the prevalence of disparities in PANE1 MiHA and its effect on graft versus host disease (GvHD) and graft versus leukemia (GvL). A sequence- specific primer (SSP) approach was used to determine the immunogenic (PANER) and non-immunogenic (PANES) alleles of PANE1 gene. Student t- test, Mann-Whitney U test, ANOVA F-test, and Kruskal-Wallis H tests were used to determine the significance of the difference for quantitative data. Relations of qualitative data were determined using Chi-square test. Survival analysis was done using Kaplan-Meier method to determine overall survival (OS). -

Influence of Chromosome 6 Deletion on the Expressional Characteristics in Humans
MOJ Biology and Medicine Mini Review Open Access Influence of chromosome 6 deletion on the expressional characteristics in humans Abstract Volume 6 Issue 1 - 2021 Deletion of chromosomal fragments bring about the devastating disease onset which Taskina Murshed, Rashed Noor is apparently almost incurable. Such deletions usually arise denovo as well as from Department of Life Sciences (DLS), School of Environment and the unstable chromosomal translocations, which in turn affect the terminal part of a Life Sciences (SELS), Independent University, Bangladesh chromosome or an interstitial region, making a large number of genes malfunctioning with the onset of various complications. Chromosome 6q deletion happens due to a missing Correspondence: Rashed Noor, Associate Professor, copy of the genetic material existing within the long arm (q) of the chromosome, which Department of Life Sciences (DLS), School of Environment and causes the loss of multiple necessary genes ending up with an array of physical and mental Life Sciences (SELS), Independent University, Bangladesh (IUB), abnormalities. current review discussed the background of such deleterious effects along Plot 16, Block B, Bashundhara, Dhaka 1229, Bangladesh, with some examples, and depicted on the possible mechanisms; i.e., the genetic insights of Tel +8801749401451, Email chromosomal breakpoint based on the information gathered from the previous literature which could be useful in perspective of the surveillance of genetic disease as well as for the Received: December 11, 2020 | Published: