The Genetical Society of Great Britain
Total Page:16
File Type:pdf, Size:1020Kb
Recommended publications
-

Screening and Identification of Key Biomarkers in Clear Cell Renal Cell Carcinoma Based on Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Screening and identification of key biomarkers in clear cell renal cell carcinoma based on bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignancy of the urinary system. The pathogenesis and effective diagnosis of ccRCC have become popular topics for research in the previous decade. In the current study, an integrated bioinformatics analysis was performed to identify core genes associated in ccRCC. An expression dataset (GSE105261) was downloaded from the Gene Expression Omnibus database, and included 26 ccRCC and 9 normal kideny samples. Assessment of the microarray dataset led to the recognition of differentially expressed genes (DEGs), which was subsequently used for pathway and gene ontology (GO) enrichment analysis. -

SPINT1) by Transcription Published: Xx Xx Xxxx Factor CDX2 E
www.nature.com/scientificreports OPEN Intestinal regulation of suppression of tumorigenicity 14 (ST14) and serine peptidase inhibitor, Kunitz Received: 5 April 2018 Accepted: 23 July 2018 type -1 (SPINT1) by transcription Published: xx xx xxxx factor CDX2 E. Thomas Danielsen 1,2, Anders Krüger Olsen2, Mehmet Coskun3, Annika W. Nonboe2, Sylvester Larsen 1,4, Katja Dahlgaard1, Eric Paul Bennett5, Cathy Mitchelmore1, Lotte Katrine Vogel2 & Jesper Thorvald Troelsen 1 The type II membrane-anchored serine protease, matriptase, encoded by suppression of tumorgenicity-14 (ST14) regulates the integrity of the intestinal epithelial barrier in concert with its inhibitor, HAI-1 encoded by serine peptidase inhibitor, Kunitz type -1 (SPINT1). The balance of the protease/inhibitor gene expression ratio is vital in preventing the oncogenic potential of matriptase. The intestinal cell lineage is regulated by a transcriptional regulatory network where the tumor suppressor, Caudal homeobox 2 (CDX2) is considered to be an intestinal master transcription factor. In this study, we show that CDX2 has a dual function in regulating both ST14 and SPINT1, gene expression in intestinal cells. We fnd that CDX2 is not required for the basal ST14 and SPINT1 gene expression; however changes in CDX2 expression afects the ST14/SPINT1 mRNA ratio. Exploring CDX2 ChIP-seq data from intestinal cell lines, we identifed genomic CDX2-enriched enhancer elements for both ST14 and SPINT1, which regulate their corresponding gene promoter activity. We show that CDX2 displays both repressive and enhancing regulatory abilities in a cell specifc manner. Together, these data reveal new insight into transcriptional mechanisms controlling the intestinal matriptase/inhibitor balance. -

SARS-Cov-2 Entry Protein TMPRSS2 and Its Homologue, TMPRSS4
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.26.441280; this version posted April 26, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 SARS-CoV-2 Entry Protein TMPRSS2 and Its 2 Homologue, TMPRSS4 Adopts Structural Fold Similar 3 to Blood Coagulation and Complement Pathway 4 Related Proteins ∗,a ∗∗,b b 5 Vijaykumar Yogesh Muley , Amit Singh , Karl Gruber , Alfredo ∗,a 6 Varela-Echavarría a 7 Instituto de Neurobiología, Universidad Nacional Autónoma de México, Querétaro, México b 8 Institute of Molecular Biosciences, University of Graz, Graz, Austria 9 Abstract The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) utilizes TMPRSS2 receptor to enter target human cells and subsequently causes coron- avirus disease 19 (COVID-19). TMPRSS2 belongs to the type II serine proteases of subfamily TMPRSS, which is characterized by the presence of the serine- protease domain. TMPRSS4 is another TMPRSS member, which has a domain architecture similar to TMPRSS2. TMPRSS2 and TMPRSS4 have been shown to be involved in SARS-CoV-2 infection. However, their normal physiological roles have not been explored in detail. In this study, we analyzed the amino acid sequences and predicted 3D structures of TMPRSS2 and TMPRSS4 to under- stand their functional aspects at the protein domain level. Our results suggest that these proteins are likely to have common functions based on their conserved domain organization. -

ST14 (NM 021978) Human 3' UTR Clone – SC207486 | Origene
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for SC207486 ST14 (NM_021978) Human 3' UTR Clone Product data: Product Type: 3' UTR Clones Product Name: ST14 (NM_021978) Human 3' UTR Clone Vector: pMirTarget (PS100062) Symbol: ST14 Synonyms: ARCI11; CAP3; HAI; MT-SP1; MTSP1; PRSS14; SNC19; TADG15; TMPRSS14 ACCN: NM_021978 Insert Size: 569 bp Insert Sequence: >SC207486 3’UTR clone of NM_021978 The sequence shown below is from the reference sequence of NM_021978. The complete sequence of this clone may contain minor differences, such as SNPs. Blue=Stop Codon Red=Cloning site GGCAAGTTGGACGCCCGCAAGATCCGCGAGATTCTCATTAAGGCCAAGAAGGGCGGAAAGATCGCCGTG TAACAATTGGCAGAGCTCAGAATTCAAGCGATCGCC GACTGGATCAAAGAGAACACTGGGGTATAGGGGCCGGGGCCACCCAAATGTGTACACCTGCGGGGCCAC CCATCGTCCACCCCAGTGTGCACGCCTGCAGGCTGGAGACTGGACCGCTGACTGCACCAGCGCCCCCAG AACATACACTGTGAACTCAATCTCCAGGGCTCCAAATCTGCCTAGAAAACCTCTCGCTTCCTCAGCCTC CAAAGTGGAGCTGGGAGGTAGAAGGGGAGGACACTGGTGGTTCTACTGACCCAACTGGGGGCAAAGGTT TGAAGACACAGCCTCCCCCGCCAGCCCCAAGCTGGGCCGAGGCGCGTTTGTGCATATCTGCCTCCCCTG TCTCTAAGGAGCAGCGGGAACGGAGCTTCGGGGCCTCCTCAGTGAAGGTGGTGGGGCTGCCGGATCTGG GCTGTGGGGCCCTTGGGCCACGCTCTTGAGGAAGCCCAGGCTCGGAGGACCCTGGAAAACAGACGGGTC TGAGACTGAAATTGTTTTACCAGCTCCCAGGGTGGACTTCAGTGTGTGTATTTGTGTAAATGAGTAAAA CATTTTATTTCTTTTTA ACGCGTAAGCGGCCGCGGCATCTAGATTCGAAGAAAATGACCGACCAAGCGACGCCCAACCTGCCATCA CGAGATTTCGATTCCACCGCCGCCTTCTATGAAAGG Restriction Sites: SgfI-MluI OTI Disclaimer: -

MECHANISMS in ENDOCRINOLOGY: Novel Genetic Causes of Short Stature
J M Wit and others Genetics of short stature 174:4 R145–R173 Review MECHANISMS IN ENDOCRINOLOGY Novel genetic causes of short stature 1 1 2 2 Jan M Wit , Wilma Oostdijk , Monique Losekoot , Hermine A van Duyvenvoorde , Correspondence Claudia A L Ruivenkamp2 and Sarina G Kant2 should be addressed to J M Wit Departments of 1Paediatrics and 2Clinical Genetics, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, Email The Netherlands [email protected] Abstract The fast technological development, particularly single nucleotide polymorphism array, array-comparative genomic hybridization, and whole exome sequencing, has led to the discovery of many novel genetic causes of growth failure. In this review we discuss a selection of these, according to a diagnostic classification centred on the epiphyseal growth plate. We successively discuss disorders in hormone signalling, paracrine factors, matrix molecules, intracellular pathways, and fundamental cellular processes, followed by chromosomal aberrations including copy number variants (CNVs) and imprinting disorders associated with short stature. Many novel causes of GH deficiency (GHD) as part of combined pituitary hormone deficiency have been uncovered. The most frequent genetic causes of isolated GHD are GH1 and GHRHR defects, but several novel causes have recently been found, such as GHSR, RNPC3, and IFT172 mutations. Besides well-defined causes of GH insensitivity (GHR, STAT5B, IGFALS, IGF1 defects), disorders of NFkB signalling, STAT3 and IGF2 have recently been discovered. Heterozygous IGF1R defects are a relatively frequent cause of prenatal and postnatal growth retardation. TRHA mutations cause a syndromic form of short stature with elevated T3/T4 ratio. Disorders of signalling of various paracrine factors (FGFs, BMPs, WNTs, PTHrP/IHH, and CNP/NPR2) or genetic defects affecting cartilage extracellular matrix usually cause disproportionate short stature. -

Chromosome 6 Abnormalities in Ovarian Surface Epithelial Tumors of Borderline Malignancy Suggest a Genetic Continuum in the Progression Model of Ovarian Neoplasms1
3404 Vol. 7, 3404–3409, November 2001 Clinical Cancer Research Chromosome 6 Abnormalities in Ovarian Surface Epithelial Tumors of Borderline Malignancy Suggest a Genetic Continuum in the Progression Model of Ovarian Neoplasms1 Maria Grazia Tibiletti, Barbara Bernasconi, Conclusion: Alterations in the above-mentioned inter- Daniela Furlan, Paola Bressan, Roberta Cerutti, val are a common finding in advanced ovarian carcinomas but also in benign ovarian cysts, implying that some tumors Carla Facco, Massimo Franchi, Cristina Riva, of borderline malignancy may arise from benign tumors and Raffaella Cinquetti, Carlo Capella, and that malignant ones may evolve from tumors of borderline Roberto Taramelli2 malignancy. Genes located in 6q27 seem to be crucial for this Laboratorio di Anatomia Patologica Ospedale di Circolo, Fondazione mechanism of early events in ovarian tumorigenesis. Macchi, 21100 Varese [M. G. T., C. F., C. R.]; and Dipartimento di Scienze Cliniche e Biologiche [B. B., D. F., P. B., R. Ce., C. C.], INTRODUCTION Clinica Ostetrica e Ginecologica [M. F.], and Dipartimento di Biologia Strutturale e Funzionale [R. Ci., R. T.], Universita` Ovarian cancer represents the most lethal of the gyneco- dell’Insubria, 21100 Varese, Italy logical tumors, and its pathogenesis is poorly understood. More than 80% of malignant tumors of the ovary are of epithelial origin and arise from the surface epithelium (1). Ovarian surface ABSTRACT epithelial tumors are divided into three histological subtypes: Purpose: We used conventional cytogenetics, molecular benign cystoadenoma, TBM,3 and AC, which are related to a cytogenetics, and molecular genetic analyses to study the different biological behavior (1). The molecular events under- pattern of allelic loss on chromosome 6q in a cohort of lying the development of ovarian tumorigenesis are still largely borderline epithelial ovarian tumors. -
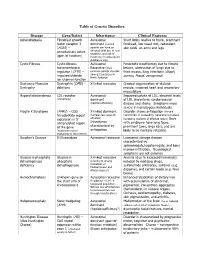
Table of Genetic Disorders Disease Gene/Defect Inheritance Clinical
Table of Genetic Disorders Disease Gene/Defect Inheritance Clinical Features Achondroplasia Fibroblast growth Autosomal Short limbs relative to trunk, prominent factor receptor 3 dominant (normal forehead, low nasal root, redundant (FGR3) – parents can have an skin folds on arms and legs constitutively active affected child due to new mutation, and risk of (gain of function) recurrence in subsequent children is low) Cystic Fibrosis Cystic fibrosis Autosomal Pancreatic insufficiency due to fibrotic transmembrane Recessive (most lesions, obstruction of lungs due to regulator (CFTR) – common genetic disorder thick mucus, lung infections (Staph, impaired chloride among Caucasians in aureus, Pseud. aeruginosa) North America) ion channel function Duchenne Muscular Dystrophin (DMD) - X-linked recessive Gradual degeneration of skeletal Dystrophy deletions muscle, impaired heart and respiratory musculature Hypercholesterolemia LDL receptor Autosomal Impaired uptake of LDL, elevated levels (commonly) dominant of LDL cholesterol, cardiovascular (haploinsufficiency) disease and stroke. Symptoms more severe in homozygous individuals Fragile X Syndrome (FMR1) – CGG X-linked dominant Disorder shows anticipation (female trinucleotide repeat (females less severely transmitters in succeeding generations produce expansion in 5’ affected) increasing numbers of affected males) Boys untranslated region Inheritance with syndrome have long faces, of the gene characterized by prominent jaws, large ears, and are (expansion occurs anticipation likely to be mentally retarded. exclusively in the mother) Gaucher’s Disease Β-Glucosidase Autosomal recessive Lysosomal storage disease characterized by splenomegaly,hepatomegaly, and bone marrow infiltration. Neurological symptoms are not common Glucose 6-phosphate Glucose 6- X-linked recessive Anemia (due to increased hemolysis) dehydrogenase phosphate (prominent among induced by oxidizing drugs, deficiency dehydrogenase individuals of sulfonamide antibiotics, sulfones (e.g. -

Molecular Cytogenetic Characterisation of a Novel De Novo Ring
Pace et al. Molecular Cytogenetics (2017) 10:9 DOI 10.1186/s13039-017-0311-y CASEREPORT Open Access Molecular cytogenetic characterisation of a novel de novo ring chromosome 6 involving a terminal 6p deletion and terminal 6q duplication in the different arms of the same chromosome Nikolai Paul Pace1, Frideriki Maggouta2, Melissa Twigden2 and Isabella Borg1,3,4* Abstract Background: Ring chromosome 6 is a rare sporadic chromosomal abnormality, associated with extreme variability in clinical phenotypes. Most ring chromosomes are known to have deletions on one or both chromosomal arms. Here, we report an atypical and unique ring chromosome 6 involving both a distal deletion and a distal duplication on the different arms of the same chromosome. Case presentation: In a patient with intellectual disability, short stature, microcephaly, facial dysmorphology, congenital heart defects and renovascular disease, a ring chromosome 6 was characterised using array-CGH and dual-colour FISH. The de-novo ring chromosome 6 involved a 1.8 Mb terminal deletion in the distal short arm and a 2.5 Mb duplication in the distal long arm of the same chromosome 6. This results in monosomy for the region 6pter to 6p25.3 and trisomy for the region 6q27 to 6qter. Analysis of genes in these chromosomal regions suggests that haploinsufficiency for FOXC1 and GMDS genes accounts for the cardiac and neurodevelopmental phenotypes in the proband. The ring chromosome 6 reported here is atypical as it involves a unique duplication of the distal long arm. Furthermore, the presence of renovascular disease is also a unique feature identified in this patient. -

WNT16 Is a New Marker of Senescence
Table S1. A. Complete list of 177 genes overexpressed in replicative senescence Value Gene Description UniGene RefSeq 2.440 WNT16 wingless-type MMTV integration site family, member 16 (WNT16), transcript variant 2, mRNA. Hs.272375 NM_016087 2.355 MMP10 matrix metallopeptidase 10 (stromelysin 2) (MMP10), mRNA. Hs.2258 NM_002425 2.344 MMP3 matrix metallopeptidase 3 (stromelysin 1, progelatinase) (MMP3), mRNA. Hs.375129 NM_002422 2.300 HIST1H2AC Histone cluster 1, H2ac Hs.484950 2.134 CLDN1 claudin 1 (CLDN1), mRNA. Hs.439060 NM_021101 2.119 TSPAN13 tetraspanin 13 (TSPAN13), mRNA. Hs.364544 NM_014399 2.112 HIST2H2BE histone cluster 2, H2be (HIST2H2BE), mRNA. Hs.2178 NM_003528 2.070 HIST2H2BE histone cluster 2, H2be (HIST2H2BE), mRNA. Hs.2178 NM_003528 2.026 DCBLD2 discoidin, CUB and LCCL domain containing 2 (DCBLD2), mRNA. Hs.203691 NM_080927 2.007 SERPINB2 serpin peptidase inhibitor, clade B (ovalbumin), member 2 (SERPINB2), mRNA. Hs.594481 NM_002575 2.004 HIST2H2BE histone cluster 2, H2be (HIST2H2BE), mRNA. Hs.2178 NM_003528 1.989 OBFC2A Oligonucleotide/oligosaccharide-binding fold containing 2A Hs.591610 1.962 HIST2H2BE histone cluster 2, H2be (HIST2H2BE), mRNA. Hs.2178 NM_003528 1.947 PLCB4 phospholipase C, beta 4 (PLCB4), transcript variant 2, mRNA. Hs.472101 NM_182797 1.934 PLCB4 phospholipase C, beta 4 (PLCB4), transcript variant 1, mRNA. Hs.472101 NM_000933 1.933 KRTAP1-5 keratin associated protein 1-5 (KRTAP1-5), mRNA. Hs.534499 NM_031957 1.894 HIST2H2BE histone cluster 2, H2be (HIST2H2BE), mRNA. Hs.2178 NM_003528 1.884 CYTL1 cytokine-like 1 (CYTL1), mRNA. Hs.13872 NM_018659 tumor necrosis factor receptor superfamily, member 10d, decoy with truncated death domain (TNFRSF10D), 1.848 TNFRSF10D Hs.213467 NM_003840 mRNA. -

Isolation of Chromosome-Specific Ests by Microdissection-Mediated Cdna Capture Edgardo Gracia, 1-3 Michael E
Downloaded from genome.cshlp.org on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press RESEARCH Isolation of Chromosome-Specific ESTs by Microdissection-Mediated cDNA Capture Edgardo Gracia, 1-3 Michael E. Ray, 1-3 Mihael H. Polymeropoulos, 4 Anindya Dehejia, 4 Paul S. Meltzer, 3 and Jeffrey M. Trent 3's 1Department of Human Genetics, The University of Michigan Medical School, Medical Science II M4708, Ann Arbor, Michigan 48109; 3Laboratory of Cancer Genetics, 4 Laboratory of Genetic Disease Research, National Center for Human Genome Research, National Institutes of Health, Bethesda, Maryland 20892 Despite dramatic advances in the identification of human expressed sequence tags (ESTs), techniques that facilitate isolation of chromosome or chromosome band-specific ESTs would be of considerable value. This report demonstrates the feasibility of identifying chromosome-specific ESTs following microdissection of a single-copy chromosome region. For this study, a reduced complexity cDNA library was linkered and hybridized to normal human metaphase chromosomes. After stringency washes, the entire long arm of chromosome 6 (6q) was microdissected. Following PCR amplification using linker-specific primers, captured cDNAs were subcloned and 187 individual clones picked at random. These 187 clones were then sorted by filter cross-hybridization into 34 unique groups. Of these 34 groups, 19 (56%) mapped to chromosome 6 by Southern blot. We identified three previously known genes, human cytovillin (ezrin) mapped previously to 6q25-26, human cardiac gap junction protein (connexin 43) mapped previously to 6q21-23.2 and prolyloligopeptidase, which had not been mapped previously. BLASTN identified three clone groups with homology to known ESTs and 12 representing novel cDNA sequences. -

Of Small Intestine Harboring Driver Gene Mutations: a Case Report and a Literature Review
1161 Case Report A rare multiple primary sarcomatoid carcinoma (SCA) of small intestine harboring driver gene mutations: a case report and a literature review Zhu Zhu1#, Xinyi Liu2#, Wenliang Li1, Zhengqi Wen1, Xiang Ji1, Ruize Zhou1, Xiaoyu Tuo3, Yaru Chen2, Xian Gong2, Guifeng Liu2, Yanqing Zhou2, Shifu Chen2, Lele Song2#^, Jian Huang1 1Department of Oncology, First Affiliated Hospital of Kunming Medical University, Kunming, China; 2HaploX Biotechnology, Shenzhen, China; 3Department of Pathology, First Affiliated Hospital of Kunming Medical University, Kunming, China #These authors contributed equally to this work. Correspondence to: Jian Huang. Department of Oncology, First Affiliated Hospital of Kunming Medical University, No. 295, Xichang Road, Kunming 560032, Yunnan Province, China. Email: [email protected]; Lele Song. HaploX Biotechnology, 8th floor, Auto Electric Power Building, Songpingshan Road, Nanshan District, Shenzhen 518057, Guangdong Province, China. Email: [email protected]. Abstract: Primary sarcomatoid carcinoma (SCA) is a type of rare tumor consisting of both malignant epithelial and mesenchymal components. Only 32 cases of SCA of the small bowel have been reported in the literature to date. Due to its rarity and complexity, this cancer has not been genetically studied and its diagnosis and treatment remain difficult. Here we report a 54-year-old male underwent emergency surgical resection in the small intestine due to severe obstruction and was diagnosed with multiple SCA based on postoperative pathological examination. Over 100 polypoid tumors scattered along his whole jejunum and proximal ileum. Chemotherapy (IFO+Epirubicin) was performed after surgery while the patient died two months after the surgery due to severe malnutrition. Whole-exome sequencing was performed for the tumor tissue with normal tissue as the control. -

Downloaded on 14Th December 2017 from SKCM 200E, 200 Kv, 4.5 Ma) 2 Days Before the Transplantation
Gómez-Abenza et al. Journal of Experimental & Clinical Cancer Research (2019) 38:405 https://doi.org/10.1186/s13046-019-1389-3 RESEARCH Open Access Zebrafish modeling reveals that SPINT1 regulates the aggressiveness of skin cutaneous melanoma and its crosstalk with tumor immune microenvironment Elena Gómez-Abenza1,2, Sofía Ibáñez-Molero1,2, Diana García-Moreno1,2, Inmaculada Fuentes1,2, Leonard I. Zon3,4, Maria C. Mione5, María L. Cayuela6, Chiara Gabellini1,2,7* and Victoriano Mulero1,2* Abstract Background: Skin cutaneous melanoma (SKCM) is the most lethal form of skin cancer and while incidence rates are declining for most cancers, they have been steadily rising for SKCM. Serine protease inhibitor, kunitz-type, 1 (SPINT1) is a type II transmembrane serine protease inhibitor that has been shown to be involved in the development of several types of cancer, such as squamous cell carcinoma and colorectal cancer. Methods: We used the unique advantages of the zebrafish to model the impact of Spint1a deficiency in early transformation, progression and metastatic invasion of SKCM together with in silico analysis of the occurrence and relevance of SPINT1 genetic alterations of the SKCM TCGA cohort. Results: We report here a high prevalence of SPINT1 genetic alterations in SKCM patients and their association with altered tumor immune microenvironment and poor patient survival. The zebrafish model reveals that Spint1a deficiency facilitates oncogenic transformation, regulates the tumor immune microenvironment crosstalk, accelerates the onset of SKCM and promotes metastatic invasion. Notably, Spint1a deficiency is required at both cell autonomous and non-autonomous levels to enhance invasiveness of SKCM. Conclusions: These results reveal a novel therapeutic target for SKCM.