A Dissertation Presented in Partial Fulfillment of the Requirements For
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Browsing Genes and Genomes with Ensembl
The Bioinformatics Roadshow Tórshavn, The Faroe Islands 28-29 November 2012 BROWSING GENES AND GENOMES WITH ENSEMBL EXERCISES AND ANSWERS 1 BROWSER 3 BIOMART 8 VARIATION 13 COMPARATIVE GENOMICS 18 2 Note: These exercises are based on Ensembl version 69 (October 2012). After in future a new version has gone live, version 69 will still be available at http://e69.ensembl.org/. If your answer doesn’t correspond with the given answer, please consult the instructor. ______________________________________________________________ BROWSER ______________________________________________________________ Exercise 1 – Exploring a gene (a) Find the human F9 (coagulation factor IX) gene. On which chromosome and which strand of the genome is this gene located? How many transcripts (splice variants) have been annotated for it? (b) What is the longest transcript? How long is the protein it encodes? Has this transcript been annotated automatically (by Ensembl) or manually (by Havana)? How many exons does it have? Are any of the exons completely or partially untranslated? (c) Have a look at the external references for ENST00000218099. What is the function of F9? (d) Is it possible to monitor expression of ENST00000218099 with the ILLUMINA HumanWG_6_V2 microarray? If so, can it also be used to monitor expression of the other two transcripts? (e) In which part (i.e. the N-terminal or C-terminal half) of the protein encoded by ENST00000218099 does its peptidase activity reside? (f) Have any missense variants been discovered for the protein encoded by ENST00000218099? (g) Is there a mouse orthologue predicted for the human F9 gene? (h) If you have yourself a gene of interest, explore what information Ensembl displays about it! ______________________________________________________________ Answer (a) 8 Go to the Ensembl homepage (http://www.ensembl.org/). -
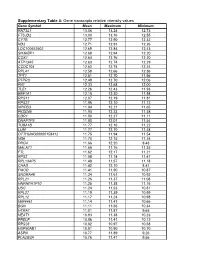
Gene Transcripts Relative Intensity Values
Supplementary Table 4: Gene transcripts relative intensity values Gene Symbol Mean Maximum Minimum RN7SL1 13.06 13.24 12.73 FTSJD2 13.00 13.16 12.55 CYTB 12.77 12.90 12.32 ND2 12.71 12.91 12.26 LOC100652902 12.69 12.84 12.43 SH3KBP1 12.68 12.94 12.20 COX1 12.64 12.76 12.20 ATP13A5 12.63 12.74 12.29 CCDC104 12.62 12.74 12.24 RPL41 12.58 12.66 12.36 TPT1 12.51 12.70 11.96 PTPRO 12.49 12.70 12.06 FN1 12.33 12.68 12.00 TLE1 12.26 12.43 11.93 EEF1A1 12.15 12.30 11.98 RPS11 12.07 12.19 11.81 RPS27 11.96 12.10 11.72 NPIPB3 11.94 12.21 11.65 FKSG49 11.94 12.23 11.58 CDR1 11.90 12.27 11.11 DNAPTP3 11.80 12.07 11.54 TUBA1B 11.77 12.16 11.22 LUM 11.77 12.10 11.48 OTTHUMG00000158412 11.75 11.94 11.54 ND6 11.70 12.15 11.24 PRG4 11.66 12.20 9.43 MALAT1 11.65 11.76 11.35 FTL 11.62 12.17 11.21 RPS2 11.58 11.74 11.47 RPL13AP5 11.48 11.57 11.18 CHAD 11.42 12.10 8.41 FMOD 11.41 11.90 10.87 SNORA48 11.34 11.61 10.92 RPL21 11.26 11.37 11.08 HNRNPA1P10 11.25 11.38 11.16 UBC 11.24 11.53 10.81 RPL27 11.19 11.39 10.99 RPL12 11.17 11.24 10.98 MIR4461 11.14 11.47 10.66 BGN 11.11 11.66 10.34 HTRA1 11.01 11.87 9.65 NEAT1 10.93 11.38 10.23 PRELP 10.86 11.41 10.12 RPS28 10.82 10.97 10.58 HSP90AB1 10.81 10.90 10.70 ASPN 10.77 11.89 9.26 PLA2G2A 10.76 11.47 9.56 H3F3A 10.69 10.86 10.41 EEF1G 10.69 10.94 10.11 C6orf48 10.64 11.02 10.35 UBA52 10.58 10.75 10.08 CTGF 10.57 11.33 9.66 MGP 10.57 11.22 10.09 YBX1 10.56 10.79 10.01 MT1X 10.56 11.89 9.88 NPC2 10.55 10.80 10.12 LAPTM4A 10.51 10.78 10.18 ITM2B 10.51 10.68 10.34 IBSP 10.50 11.50 7.17 NPIPB5 10.50 10.73 10.26 TUBA1A -

This Item Is the Archived Peer-Reviewed Author-Version Of
This item is the archived peer-reviewed author-version of: Deiodinase knockdown affects zebrafish eye development at the level of gene expression, morphology and function Reference: Houbrechts Anne M., Vergauwen Lucia, Bagci Enise, Van houcke Jolien, Heijlen Marjolein, Kulemeka Bernard, Hyde David R., Knapen Dries, Darras Veerle M..- Deiodinase knockdown affects zebrafish eye development at the level of gene expression, morphology and function Molecular and cellular endocrinology - ISSN 0303-7207 - 424(2016), p. 81-93 Full text (Publishers DOI): http://dx.doi.org/doi:10.1016/j.mce.2016.01.018 To cite this reference: http://hdl.handle.net/10067/1314550151162165141 Institutional repository IRUA 1 Deiodinase knockdown affects zebrafish eye development at the level 2 of gene expression, morphology and function. 3 Anne M. Houbrechts1*, Lucia Vergauwen2*, Enise Bagci2,3, Jolien Van houcke1, Marjolein 4 Heijlen1, Bernard Kulemeka4, David R. Hyde4, Dries Knapen2, Veerle M. Darras1 5 1 Laboratory of Comparative Endocrinology, Department of Biology, Division of 6 Animal Physiology and Neurobiology, KU Leuven, 3000 Leuven, Belgium 7 2 Zebrafishlab, Veterinary Physiology and Biochemistry, Department of Veterinary 8 Sciences, University of Antwerp, Universiteitsplein 1, 2610 Wilrijk, Belgium 9 3 Systemic Physiological & Ecotoxicological Research, Department of Biology, 10 University of Antwerp, Universiteitsplein 1, 2610 Wilrijk, Belgium 11 4 Department of Biological Sciences and Center for Zebrafish Research, University of 12 Notre Dame, Notre Dame, -

A Brain-Derived Neurotrophic Factor
www.nature.com/scientificreports OPEN A Brain-Derived Neurotrophic Factor Mimetic Is Sufcient to Restore Cone Photoreceptor Visual Received: 17 January 2017 Accepted: 25 August 2017 Function in an Inherited Blindness Published: xx xx xxxx Model Conor Daly1, Lisa Shine1, Theresa Hefernan1, Sudhakar Deeti1, Alison L. Reynolds 1, John J. O’Connor1, Eugène T. Dillon 1,2, David J. Dufy 3,5, Walter Kolch 1,3,4, Gerard Cagney1 & Breandán N. Kennedy 1 Controversially, histone deacetylase inhibitors (HDACi) are in clinical trial for the treatment of inherited retinal degeneration. Utilizing the zebrafsh dyeucd6 model, we determined if treatment with HDACi can rescue cone photoreceptor-mediated visual function. dye exhibit defective visual behaviour and retinal morphology including ciliary marginal zone (CMZ) cell death and decreased photoreceptor outer segment (OS) length, as well as gross morphological defects including hypopigmentation and pericardial oedema. HDACi treatment of dye results in signifcantly improved optokinetic (OKR) (~43 fold, p < 0.001) and visualmotor (VMR) (~3 fold, p < 0.05) responses. HDACi treatment rescued gross morphological defects and reduced CMZ cell death by 80%. Proteomic analysis of dye eye extracts suggested BDNF-TrkB and Akt signaling as mediators of HDACi rescue in our dataset. Co- treatment with the TrkB antagonist ANA-12 blocked HDACi rescue of visual function and associated Akt phosphorylation. Notably, sole treatment with a BDNF mimetic, 7,8-dihydroxyfavone hydrate, signifcantly rescued dye visual function (~58 fold increase in OKR, p < 0.001, ~3 fold increase in VMR, p < 0.05). In summary, HDACi and a BDNF mimetic are sufcient to rescue retinal cell death and visual function in a vertebrate model of inherited blindness. -
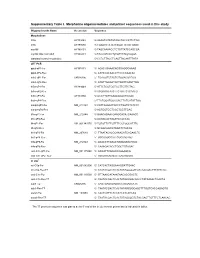
Supplementary Table I. Morpholino Oligonucleotides and Primer Sequences Used in This Study
Supplementary Table I. Morpholino oligonucleotides and primer sequences used in this study Oligonucleotide Name Accession Sequence Morpholinos tlr5a AY389449 5'-AAAGTGTATGTAGCTGCCATTCTGG tlr5b AY389450 5'-TGAATGTATATCCCATTCTGTGAGC myd88 AY388401 5'-TAGCAAAACCTCTGTTATCCAGCGA myd88 5bp mismatch AY388401 5'-TAcCAtAACCTgTGTTATCgAGgGA standard control morpholino 5'-CCTCTTACCTCAGTTACAATTTATA qRT-PCR ppial-qP1-Fw AY391451 5’- ACACTGAAACACGGAGGCAAAG ppial-qP2-Rev 5’- CATCCACAACCTTCCCGAACAC irak3-qP1-Fw CK026195 5’- TGAGGTCTACTGTGGACGATGG irak3-qP2-Rev 5’- ATGTTAGGATGCTGGTTGAGTTGG tlr5a-qP1-Fw AY389449 5’-ATTCTGGTGGTGCTTGTTGTAG tlr5a-qP2-Rev 5’-ACGAGGTAACTTCTGTTCTCAATG tlr5b-qP3-Fw AY389450 5’-GCGTTGTTGAAGAGGCTGGAC tlr5b-qP4-Rev 5’-TTCTGGATGGCCACTTCTCATATTGG mmp9-qP3-Fw NM_213123 5’-CATTAAAGATGCCCTGATGTATCCC mmp9-qP4-Rev 5’-AGTGGTGGTCCGTGGTTGAG il1b-qP1-Fw NM_212844 5’-GAACAGAATGAAGCACATCAAACC il1b-qP2-Rev 5’-ACGGCACTGAATCCACCAC il8-qP1-Fw XM_001342570 5’-TGTGTTATTGTTTTCCTGGCATTTC il8-qP2-Rev 5’-GCGACAGCGTGGATCTACAG ifn1-qP3-Fw NM_207640 5’- TTAATACACGCAAAGATGAGAACTC ifn1-qP4-Rev 5’- GCCAAGCCATTCGCAAGTAG tnfa-qP5-Fw NM_212829 5’- AGACCTTAGACTGGAGAGATGAC tnfa-qP6-Rev 5’- CAAAGACACCTGGCTGTAGAC cxcl-C1c-qP1-Fw NM_001115060 5’- GGCATTCACACCCAAAGCG cxcl-C1c-qP2_Rev 5’- GCGAGCACGATTCACGAGAG * In situ ccl-C5a-Fw NM_001082906 5’- CATCACTAGGAAAGGATTGAAC ccl-C5a-Rev-T7 5’- TAATACGACTCACTATAGGGGATGTCAAAGACTTTATTCAC cxcl-C1c-Fw NM_001115060 5’- GTTAAACATAAATAACACCGACTC cxcl-C1c-Rev-T7 5’- TAATACGACTCACTATAGGGACACCCTATAAAACTGAGTA irak3-Fw CK026195 5’- CAGTGAGAGAGGCATGAAACATC -

Supplementary Data
SUPPLEMENTARY METHODS 1) Characterisation of OCCC cell line gene expression profiles using Prediction Analysis for Microarrays (PAM) The ovarian cancer dataset from Hendrix et al (25) was used to predict the phenotypes of the cell lines used in this study. Hendrix et al (25) analysed a series of 103 ovarian samples using the Affymetrix U133A array platform (GEO: GSE6008). This dataset comprises clear cell (n=8), endometrioid (n=37), mucinous (n=13) and serous epithelial (n=41) primary ovarian carcinomas and samples from 4 normal ovaries. To build the predictor, the Prediction Analysis of Microarrays (PAM) package in R environment was employed (http://rss.acs.unt.edu/Rdoc/library/pamr/html/00Index.html). When more than one probe described the expression of a given gene, we used the probe with the highest median absolute deviation across the samples. The dataset from Hendrix et al. (25) and the dataset of OCCC cell lines described in this manuscript were then overlaid on the basis of 11536 common unique HGNC gene symbols. Only the 99 primary ovarian cancers samples and the four normal ovary samples were used to build the predictor. Following leave one out cross-validation, a predictor based upon 126 genes was able to identify correctly the four distinct phenotypes of primary ovarian tumour samples with a misclassification rate of 18.3%. This predictor was subsequently applied to the expression data from the 12 OCCC cell lines to determine the likeliest phenotype of the OCCC cell lines compared to primary ovarian cancers. Posterior probabilities were estimated for each cell line in comparison to the following phenotypes: clear cell, endometrioid, mucinous and serous epithelial. -

Autophagy Supports Color Vision
BASIC RESEARCH PAPER Autophagy 11:10, 1821--1832; October 2015; © 2015 Taylor & Francis Group, LLC Autophagy supports color vision Zhenqing Zhou, Frans Vinberg, Frank Schottler, Teresa A Doggett, Vladimir J Kefalov, and Thomas A Ferguson* Department of Ophthalmology and Visual Sciences; Washington University in St. Louis; School of Medicine; St. Louis, MO USA Keywords: autophagy, color vision, cones, degeneration, photoreceptors, retina Abbreviations: AMD, age-related macular degeneration; AMPK, AMP-activated protein kinase; ATG5, autophagy-related 5; CIS, cone inner segment; COS, cone outer segment; ERG, electroretinogram; GFP, green fluorescent protein; LC3B, microtubule-associ- ated protein 1 light chain 3 b; LCA, Leber’s congenital amaurosis; L/D, 12 h light/12 h dark; ONL, outer nuclear layer; OS, outer segment; p-AMPK, phosphorylated AMPK; PARK2/Parkin, parkin RBR ubiquitin protein ligase; PFA, paraformaldehyde; PINK1, PTEN-induced putative kinase 1; PNA, peanut agglutinin; ROS, reactive oxygen species; RP, retinitis pigmentosa; TEM, transmis- sion electron microscopy; TOMM20/TOM20, translocase of outer mitochondrial membrane 20 homolog (yeast) Cones comprise only a small portion of the photoreceptors in mammalian retinas. However, cones are vital for color vision and visual perception, and their loss severely diminishes the quality of life for patients with retinal degenerative diseases. Cones function in bright light and have higher demand for energy than rods; yet, the mechanisms that support the energy requirements of cones are poorly understood. One such pathway that potentially could sustain cones under basal and stress conditions is macroautophagy. We addressed the role of macroautophagy in cones by examining how the genetic block of this pathway affects the structural integrity, survival, and function of these neurons. -

The Vertebrate Ancestral Repertoire of Visual Opsins, Transducin Alpha Subunits and Oxytocin/ Vasopressin Receptors Was Establis
Lagman et al. BMC Evolutionary Biology 2013, 13:238 http://www.biomedcentral.com/1471-2148/13/238 RESEARCH ARTICLE Open Access The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/ vasopressin receptors was established by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications David Lagman1†, Daniel Ocampo Daza1†, Jenny Widmark1,XesúsMAbalo1, Görel Sundström1,2 and Dan Larhammar1* Abstract Background: Vertebrate color vision is dependent on four major color opsin subtypes: RH2 (green opsin), SWS1 (ultraviolet opsin), SWS2 (blue opsin), and LWS (red opsin). Together with the dim-light receptor rhodopsin (RH1), these form the family of vertebrate visual opsins. Vertebrate genomes contain many multi-membered gene families that can largely be explained by the two rounds of whole genome duplication (WGD) in the vertebrate ancestor (2R) followed by a third round in the teleost ancestor (3R). Related chromosome regions resulting from WGD or block duplications are said to form a paralogon. We describe here a paralogon containing the genes for visual opsins, the G-protein alpha subunit families for transducin (GNAT) and adenylyl cyclase inhibition (GNAI), the oxytocin and vasopressin receptors (OT/VP-R), and the L-type voltage-gated calcium channels (CACNA1-L). Results: Sequence-based phylogenies and analyses of conserved synteny show that the above-mentioned gene families, and many neighboring gene families, expanded in the early vertebrate WGDs. This allows us to deduce the following evolutionary scenario: The vertebrate ancestor had a chromosome containing the genes for two visual opsins, one GNAT, one GNAI, two OT/VP-Rs and one CACNA1-L gene. -

Us 2018 / 0305689 A1
US 20180305689A1 ( 19 ) United States (12 ) Patent Application Publication ( 10) Pub . No. : US 2018 /0305689 A1 Sætrom et al. ( 43 ) Pub . Date: Oct. 25 , 2018 ( 54 ) SARNA COMPOSITIONS AND METHODS OF plication No . 62 /150 , 895 , filed on Apr. 22 , 2015 , USE provisional application No . 62/ 150 ,904 , filed on Apr. 22 , 2015 , provisional application No. 62 / 150 , 908 , (71 ) Applicant: MINA THERAPEUTICS LIMITED , filed on Apr. 22 , 2015 , provisional application No. LONDON (GB ) 62 / 150 , 900 , filed on Apr. 22 , 2015 . (72 ) Inventors : Pål Sætrom , Trondheim (NO ) ; Endre Publication Classification Bakken Stovner , Trondheim (NO ) (51 ) Int . CI. C12N 15 / 113 (2006 .01 ) (21 ) Appl. No. : 15 /568 , 046 (52 ) U . S . CI. (22 ) PCT Filed : Apr. 21 , 2016 CPC .. .. .. C12N 15 / 113 ( 2013 .01 ) ; C12N 2310 / 34 ( 2013. 01 ) ; C12N 2310 /14 (2013 . 01 ) ; C12N ( 86 ) PCT No .: PCT/ GB2016 /051116 2310 / 11 (2013 .01 ) $ 371 ( c ) ( 1 ) , ( 2 ) Date : Oct . 20 , 2017 (57 ) ABSTRACT The invention relates to oligonucleotides , e . g . , saRNAS Related U . S . Application Data useful in upregulating the expression of a target gene and (60 ) Provisional application No . 62 / 150 ,892 , filed on Apr. therapeutic compositions comprising such oligonucleotides . 22 , 2015 , provisional application No . 62 / 150 ,893 , Methods of using the oligonucleotides and the therapeutic filed on Apr. 22 , 2015 , provisional application No . compositions are also provided . 62 / 150 ,897 , filed on Apr. 22 , 2015 , provisional ap Specification includes a Sequence Listing . SARNA sense strand (Fessenger 3 ' SARNA antisense strand (Guide ) Mathew, Si Target antisense RNA transcript, e . g . NAT Target Coding strand Gene Transcription start site ( T55 ) TY{ { ? ? Targeted Target transcript , e . -
Università Degli Studi Di Ferrara
Università degli Studi di Ferrara DOTTORATO DI RICERCA IN BIOLOGIA EVOLUZIONISTICA E AMBIENTALE CICLO XXVII COORDINATORE Prof. GUIDO BARBUJANI Molecular and Bioinformatic Analysis of the Circadian Clock in Phreatichthys andruzzii Settore Scientifico Disciplinare BIO/05 Dottorando Tutore Dott. NEGRINI PIETRO Prof. BERTOLUCCI CRISTIANO Anni 2012/2014 2 TABLE OF CONTENTS Page Abstract (English)...…………………………………………………............................7 Abstract (Italiano)...........................................................................................................9 I. Introduction………………………………………………....................................11 1.1 General aspects of the circadian clock……………………………………….13 1.1.1 Circadian pacemakers………………………..........................................................13 1.1.2 Molecular clock mechanism…………………………………………………….…15 1.1.3 Light input pathways……………………………………………………...…….…19 1.1.4 Temperature input pathway………………………………………...……………..21 1.1.5 Output pathways…………………………………………………………………...22 1.2 Zebrafish as a circadian clock model system……………...………………22 1.2.1 Zebrafish circadian clock genes…………………………………………….……..24 1.2.2 Zebrafish light input pathway……………………………………………………..24 1.2.3 Zebrafish temperature input pathway…………..…………………………..……27 1.2.4 The role of the SCN and pineal gland in zebrafish…….…………………..…….27 1.2.5 Circadian clock outputs in zebrafish………………………………………….…..28 1.3 Cavefish as a new circadian clock model system………………..…….….29 1.3.1 Life in the darkness: special features of cave inhabitants………………….…....29 -
Genome of the Chinese Tree Shrew
ARTICLE Received 6 Sep 2012 | Accepted 20 Dec 2012 | Published 5 Feb 2013 DOI: 10.1038/ncomms2416 OPEN Genome of the Chinese tree shrew Yu Fan1,2,*, Zhi-Yong Huang3,*, Chang-Chang Cao3, Ce-Shi Chen1, Yuan-Xin Chen3, Ding-Ding Fan3, Jing He3, Hao-Long Hou3,LiHu3, Xin-Tian Hu1, Xuan-Ting Jiang3, Ren Lai1, Yong-Shan Lang3, Bin Liang1, Sheng-Guang Liao3, Dan Mu1,2, Yuan-Ye Ma1, Yu-Yu Niu1, Xiao-Qing Sun3, Jin-Quan Xia3, Jin Xiao3, Zhi-Qiang Xiong3, Lin Xu1, Lan Yang3, Yun Zhang1, Wei Zhao3, Xu-Dong Zhao1, Yong-Tang Zheng1, Ju-Min Zhou1, Ya-Bing Zhu3, Guo-Jie Zhang1,3,5, Jun Wang3,4,5,6 & Yong-Gang Yao1 Chinese tree shrews (Tupaia belangeri chinensis) possess many features valuable in animals used as experimental models in biomedical research. Currently, there are numerous attempts to employ tree shrews as models for a variety of human disorders: depression, myopia, hepatitis B and C virus infections, and hepatocellular carcinoma, to name a few. Here we present a publicly available annotated genome sequence for the Chinese tree shrew. Phy- logenomic analysis of the tree shrew and other mammalians highly support its close affinity to primates. By characterizing key factors and signalling pathways in nervous and immune systems, we demonstrate that tree shrews possess both shared common and unique features, and provide a genetic basis for the use of this animal as a potential model for biomedical research. 1 Key Laboratory of Animal Models and Human Disease Mechanisms of Chinese Academy of Sciences and Yunnan Province, Kunming Institute of Zoology, Kunming, Yunnan 650223, China. -

Proteomic Landscape of the Human Choroid–Retinal Pigment Epithelial Complex
Supplementary Online Content Skeie JM, Mahajan VB. Proteomic landscape of the human choroid–retinal pigment epithelial complex. JAMA Ophthalmol. Published online July 24, 2014. doi:10.1001/jamaophthalmol.2014.2065. eFigure 1. Choroid–retinal pigment epithelial (RPE) proteomic analysis pipeline. eFigure 2. Gene ontology (GO) distributions and pathway analysis of human choroid– retinal pigment epithelial (RPE) protein show tissue similarity. eMethods. Tissue collection, mass spectrometry, and analysis. eTable 1. Complete table of proteins identified in the human choroid‐RPE using LC‐ MS/MS. eTable 2. Top 50 signaling pathways in the human choroid‐RPE using MetaCore. eTable 3. Top 50 differentially expressed signaling pathways in the human choroid‐RPE using MetaCore. eTable 4. Differentially expressed proteins in the fovea, macula, and periphery of the human choroid‐RPE. eTable 5. Differentially expressed transcription proteins were identified in foveal, macular, and peripheral choroid‐RPE (p<0.05). eTable 6. Complement proteins identified in the human choroid‐RPE. eTable 7. Proteins associated with age related macular degeneration (AMD). This supplementary material has been provided by the authors to give readers additional information about their work. © 2014 American Medical Association. All rights reserved. 1 Downloaded From: https://jamanetwork.com/ on 09/25/2021 eFigure 1. Choroid–retinal pigment epithelial (RPE) proteomic analysis pipeline. A. The human choroid‐RPE was dissected into fovea, macula, and periphery samples. B. Fractions of proteins were isolated and digested. C. The peptide fragments were analyzed using multi‐dimensional LC‐MS/MS. D. X!Hunter, X!!Tandem, and OMSSA were used for peptide fragment identification. E. Proteins were further analyzed using bioinformatics.