Partitioning of Gene Expression Among Zebrafish Photoreceptor
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Browsing Genes and Genomes with Ensembl
The Bioinformatics Roadshow Tórshavn, The Faroe Islands 28-29 November 2012 BROWSING GENES AND GENOMES WITH ENSEMBL EXERCISES AND ANSWERS 1 BROWSER 3 BIOMART 8 VARIATION 13 COMPARATIVE GENOMICS 18 2 Note: These exercises are based on Ensembl version 69 (October 2012). After in future a new version has gone live, version 69 will still be available at http://e69.ensembl.org/. If your answer doesn’t correspond with the given answer, please consult the instructor. ______________________________________________________________ BROWSER ______________________________________________________________ Exercise 1 – Exploring a gene (a) Find the human F9 (coagulation factor IX) gene. On which chromosome and which strand of the genome is this gene located? How many transcripts (splice variants) have been annotated for it? (b) What is the longest transcript? How long is the protein it encodes? Has this transcript been annotated automatically (by Ensembl) or manually (by Havana)? How many exons does it have? Are any of the exons completely or partially untranslated? (c) Have a look at the external references for ENST00000218099. What is the function of F9? (d) Is it possible to monitor expression of ENST00000218099 with the ILLUMINA HumanWG_6_V2 microarray? If so, can it also be used to monitor expression of the other two transcripts? (e) In which part (i.e. the N-terminal or C-terminal half) of the protein encoded by ENST00000218099 does its peptidase activity reside? (f) Have any missense variants been discovered for the protein encoded by ENST00000218099? (g) Is there a mouse orthologue predicted for the human F9 gene? (h) If you have yourself a gene of interest, explore what information Ensembl displays about it! ______________________________________________________________ Answer (a) 8 Go to the Ensembl homepage (http://www.ensembl.org/). -

Supplemental Materials ZNF281 Enhances Cardiac Reprogramming
Supplemental Materials ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression Huanyu Zhou, Maria Gabriela Morales, Hisayuki Hashimoto, Matthew E. Dickson, Kunhua Song, Wenduo Ye, Min S. Kim, Hanspeter Niederstrasser, Zhaoning Wang, Beibei Chen, Bruce A. Posner, Rhonda Bassel-Duby and Eric N. Olson Supplemental Table 1; related to Figure 1. Supplemental Table 2; related to Figure 1. Supplemental Table 3; related to the “quantitative mRNA measurement” in Materials and Methods section. Supplemental Table 4; related to the “ChIP-seq, gene ontology and pathway analysis” and “RNA-seq” and gene ontology analysis” in Materials and Methods section. Supplemental Figure S1; related to Figure 1. Supplemental Figure S2; related to Figure 2. Supplemental Figure S3; related to Figure 3. Supplemental Figure S4; related to Figure 4. Supplemental Figure S5; related to Figure 6. Supplemental Table S1. Genes included in human retroviral ORF cDNA library. Gene Gene Gene Gene Gene Gene Gene Gene Symbol Symbol Symbol Symbol Symbol Symbol Symbol Symbol AATF BMP8A CEBPE CTNNB1 ESR2 GDF3 HOXA5 IL17D ADIPOQ BRPF1 CEBPG CUX1 ESRRA GDF6 HOXA6 IL17F ADNP BRPF3 CERS1 CX3CL1 ETS1 GIN1 HOXA7 IL18 AEBP1 BUD31 CERS2 CXCL10 ETS2 GLIS3 HOXB1 IL19 AFF4 C17ORF77 CERS4 CXCL11 ETV3 GMEB1 HOXB13 IL1A AHR C1QTNF4 CFL2 CXCL12 ETV7 GPBP1 HOXB5 IL1B AIMP1 C21ORF66 CHIA CXCL13 FAM3B GPER HOXB6 IL1F3 ALS2CR8 CBFA2T2 CIR1 CXCL14 FAM3D GPI HOXB7 IL1F5 ALX1 CBFA2T3 CITED1 CXCL16 FASLG GREM1 HOXB9 IL1F6 ARGFX CBFB CITED2 CXCL3 FBLN1 GREM2 HOXC4 IL1F7 -

Deletion of Vax1 from Gonadotropin-Releasing Hormone (Gnrh) Neurons Abolishes Gnrh Expression and Leads to Hypogonadism and Infertility
3506 • The Journal of Neuroscience, March 23, 2016 • 36(12):3506–3518 Cellular/Molecular Deletion of Vax1 from Gonadotropin-Releasing Hormone (GnRH) Neurons Abolishes GnRH Expression and Leads to Hypogonadism and Infertility Hanne M. Hoffmann,1 Crystal Trang,1 Ping Gong,1 Ikuo Kimura,2 Erica C. Pandolfi,1 and XPamela L. Mellon1 1Department of Reproductive Medicine and the Center for Reproductive Science and Medicine, University of California, San Diego, La Jolla, California 92093-0674, and 2Department of Applied Biological Science, Graduate School of Agriculture, Tokyo University of Agriculture and Technology, Fuchu-shi 183-8509, Japan Hypothalamic gonadotropin-releasing hormone (GnRH) neurons are at the apex of the hypothalamic-pituitary-gonadal axis that regu- lates mammalian fertility. Herein we demonstrate a critical role for the homeodomain transcription factor ventral anterior homeobox 1 (VAX1) in GnRH neuron maturation and show that Vax1 deletion from GnRH neurons leads to complete infertility in males and females. Specifically, global Vax1 knock-out embryos had normal numbers of GnRH neurons at 13 d of gestation, but no GnRH staining was detected by embryonic day 17. To identify the role of VAX1 specifically in GnRH neuron development, Vax1flox mice were generated and lineage tracing performed in Vax1flox/flox:GnRHcre:RosaLacZ mice. This identified VAX1 as essential for maintaining expression of Gnrh1. The absence of GnRH staining in adult Vax1flox/flox:GnRHcre mice led to delayed puberty, hypogonadism, and infertility. To address the mechanism by which VAX1 maintains Gnrh1 transcription, the capacity of VAX1 to regulate Gnrh1 transcription was evaluated in the GnRH cell lines GN11 and GT1-7. -

Lncrna Mediated Hijacking of T-Cell Hypoxia Response Pathway by Mycobacterium Tuberculosis Predicts Latent to Active Progression in Humans
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.11.037176; this version posted April 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. lncRNA Mediated Hijacking of T-cell Hypoxia Response Pathway by Mycobacterium Tuberculosis Predicts Latent to Active Progression in Humans 1# 1# 2# 1* Jyotsana Mehra , Vikram Kumar , Priyansh Srivastava , Tavpritesh Sethi 1 1 2 1* [email protected] , [email protected] , [email protected] , [email protected] 1 2 Indraprastha Institute of Information Technology, Delhi, India, Amity University, Noida, Uttar Pradesh, India # * Equal Contribution, Corresponding Author Abstract- Cytosolic functions of Long non-coding RNAs including mRNA translation masking and sponging are major regulators of biological pathways. Formation of T cell- bounded hypoxic granuloma is a host immune defense for containing infected Mtb-macrophages. Our study exploits the mechanistic pathway of Mtb-induced HIF1A silencing by the antisense lncRNA-HIF1A-AS2 in T cells. Computational analysis of in-vitro T-cell stimulation assays in progressors(n=119) versus latent(n=221) tuberculosis patients revealed the role of lncRNA mediated disruption of hypoxia adaptation pathways in progressors. We found 291 upregulated and 227 downregulated lncRNAs that were correlated at mRNA level with HIF1A and HILPDA which are major players in hypoxia response. We also report novel lncRNA-AC010655 (AC010655.4 and AC010655.2) in cis with HILPDA, both of which contain binding sites for the BARX2 transcription factor, thus indicating a mechanistic role. -

Original Article the Association Between the EPHA3 Gene Polymorphism and Non-Syndromic Cleft Lip with Or Without Palate in the Western Han Chinese Population
Int J Clin Exp Med 2020;13(9):7060-7070 www.ijcem.com /ISSN:1940-5901/IJCEM0112790 Original Article The association between the EPHA3 gene polymorphism and non-syndromic cleft lip with or without palate in the western Han Chinese population Yulan Zhou1*, Jiayu Shi2*, Bing Shi1, Zhonglin Jia1 1State Key Laboratory of Oral Diseases & National Clinical Research Center for Oral Diseases & Department of Cleft Lip and Palate, West China Hospital of Stomatology, Sichuan University, Chengdu, China; 2Division of Growth and Development and Section of Orthodontics, School of Dentistry, University of California, Los Angeles, USA. *Equal contributors. Received April 29, 2020; Accepted July 17, 2020; Epub September 15, 2020; Published September 30, 2020 Abstract: Non-syndromic cleft lip with or without palate (NSCL/P) is a common craniofacial congenital disease which results from multiple susceptibility genes and adverse environmental factors. A recent study reported that the T al- lele of rs7650466 of the EPHA3 gene is a genetic risk factor in the etiology of NSCL/P among the northern Chinese Han population. This study aimed to evaluate the association between the EPHA3 gene variations and NSCL/P in the western Han Chinese population. Here, we conducted targeted region sequencing around rs7632427 (EPHA3) among 159 unrelated NSCL/P cases from the western Han Chinese population and performed a gene-based burden analysis on the rare variations and a single variation association analysis on the common SNVs (single nucleotide variants). Then we found 438 SNVs and 120 indels in all. A burden analysis showed no statistical signifi- cance. The association analysis results suggested that the common SNV rs13094064 was associated with NSCL/P (P=1.86E-07 and OR=0.17), and rs7632427 was most closely related to NSCLO (P=7.49E-05 and OR=0.315). -

University of Dundee Molecular Screening of VAX1 Gene
University of Dundee Molecular Screening of VAX1 Gene Polymorphisms Uncovered the Genetic Heterogeneity of Non-Syndromic Orofacial Cleft in Saudi Arabian Patients Jafar Sabbagh, Heba ; Innes, Nicola; Edris Ahmed, Sherif ; Butali, Azeez; Abdulbaset Alnamnakani, Eman ; Rabah, Sari Published in: Genetic Testing and Molecular Biomarkers DOI: 10.1089/gtmb.2018.0207 Publication date: 2019 Document Version Peer reviewed version Link to publication in Discovery Research Portal Citation for published version (APA): Jafar Sabbagh, H., Innes, N., Edris Ahmed, S., Butali, A., Abdulbaset Alnamnakani, E., Rabah, S., Hamdan, M. A., Alhamlan, N., Abdulhameed, F. D., Hassan, M. H. A., Bassam Al Mahdi, H., Alamoudi, N. M., Alaki, S. M., & Mossey, P. (2019). Molecular Screening of VAX1 Gene Polymorphisms Uncovered the Genetic Heterogeneity of Non-Syndromic Orofacial Cleft in Saudi Arabian Patients. Genetic Testing and Molecular Biomarkers, 23(1), 45- 50. https://doi.org/10.1089/gtmb.2018.0207 General rights Copyright and moral rights for the publications made accessible in Discovery Research Portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from Discovery Research Portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain. • You may freely distribute the URL identifying the publication in the public portal. Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. -

Clinical and Molecular Investigation of Rare
CLINICAL AND MOLECULAR INVESTIGATION OF RARE CONGENITAL DEFECTS OF THE PALATE RIMANTE SESELGYTE A thesis submitted for the degree of Doctor of Philosophy to University College London August 2019 page Title 1 DECLARATION I, Rimante Seselgyte, confirm that the work presented in this thesis is my own. Where information has been derived from other sources, I confirm that this has been indicated in the thesis. Signed……………………….. Declaration 3 ABSTRACT Cleft palate (CP) affects around 1/1500 live births and, along with cleft lip, is one of the most common forms of birth defect. The studies presented here focus on unusual defects of the palate, especially to understand better the rarely reported but surprisingly common condition called submucous cleft palate (SMCP). The frequency and consequences of SMCP from a surgical perspective were first investigated based on the caseload of the North Thames Cleft Service at Great Ormond Street Hospital and St Andrew's Centre, Broomfield Hospital, Mid Essex Hospitals Trust. It was previously reported that up to 80% of individuals with unrepaired SMCP experience speech difficulties as a consequence of velopharyngeal insufficiency (VPI). Attempted repair of the palatal defect can sometimes give poor results, so controversies still exist about the correct choice of surgical technique to use. Over 23 years, 222 patients at The North Thames Cleft Service underwent operations to manage SMCP. Nearly half of them (42.8%) were diagnosed with 22q11.2 deletion syndrome (22q11.2 DS). The first operation was palate repair, with an exception of one case, followed by a second surgical intervention required in approximately half of the patients. -

Regulatory Interactions Between Vax1, Pax6, and Mir-7 Regionalize the Lateral Ventricular-Subventricular Zone During Post-Natal Olfactory Bulb Neurogenesis in Mice
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.24.059428; this version posted April 25, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Regulatory interactions between Vax1, Pax6, and miR-7 regionalize the lateral Ventricular-Subventricular Zone during post-natal Olfactory Bulb neurogenesis in mice Nathalie Coré 1, Andrea Erni 1, Pamela L. Mellon 2, Hanne M. Hoffmann 2, Christophe Béclin 1 and Harold Cremer 1 1. Aix Marseille University, CNRS, IBDM, Campus de Luminy, Marseille, France 2. Department of Obstetrics, Gynecology, and Reproductive Sciences and the Center for Reproductive Science and Medicine, University of California, San Diego, La Jolla, California 92093-0674, USA For correspondence: [email protected] and [email protected] Abstract Several subtypes of interneurons destined for the olfactory bulb are continuously generated after birth by neural stem cells located in the ventricular-subventricular zones of the lateral ventricles. Future neuronal identity depends on the positioning of pre-determined neural stem cells along the ventricle walls, which, in turn, depends on delimited expression domains of transcription factors and their cross regulatory interactions. However, mechanisms underlying positional identity of neural stem cells are still poorly understood. Here we show that the transcription factor Vax1 controls the production of two specific neuronal sub-types. First, it is directly necessary to generate Calbindin expressing interneurons from ventro-lateral progenitors. Second, it represses the generation of dopaminergic neurons by dorso-lateral progenitors through inhibiting Pax6 expression in the dorso-lateral wall. -

The LIM Protein Complex Establishes a Retinal Circuitry of Visual Adaptation
RESEARCH ARTICLE The LIM protein complex establishes a retinal circuitry of visual adaptation by regulating Pax6 a-enhancer activity Yeha Kim1, Soyeon Lim1†, Taejeong Ha1†, You-Hyang Song1, Young-In Sohn1, Dae-Jin Park2, Sun-Sook Paik3, Joo-ri Kim-Kaneyama4, Mi-Ryoung Song5, Amanda Leung6, Edward M Levine6, In-Beom Kim3, Yong Sook Goo2, Seung-Hee Lee1, Kyung Hwa Kang7, Jin Woo Kim1* 1Department of Biological Sciences, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, South Korea; 2Department of Physiology, Chungbuk National University School of Medicine, Cheongju, South Korea; 3Department of Anatomy, College of Medicine, The Catholic University of Korea, Seoul, South Korea; 4Department of Biochemistry, Showa University School of Medicine, Tokyo, Japan; 5Department of Life Sciences, Gwangju Institute of Science and Technology (GIST), Gwangju, South Korea; 6Department of Ophthalmology and Visual Sciences, Vanderbilt University, Nashville, United States; 7KAIST Institute of BioCentury, Daejeon, South Korea Abstract The visual responses of vertebrates are sensitive to the overall composition of retinal interneurons including amacrine cells, which tune the activity of the retinal circuitry. The expression of Paired-homeobox 6 (PAX6) is regulated by multiple cis-DNA elements including the intronic a- *For correspondence: enhancer, which is active in GABAergic amacrine cell subsets. Here, we report that the [email protected] transforming growth factor ß1-induced transcript 1 protein (Tgfb1i1) interacts with the LIM domain transcription factors Lhx3 and Isl1 to inhibit the a-enhancer in the post-natal mouse retina. †These authors contributed Tgfb1i1-/- mice show elevated a-enhancer activity leading to overproduction of Pax6DPD isoform equally to this work that supports the GABAergic amacrine cell fate maintenance. -

Identification of Shared and Unique Gene Families Associated with Oral
International Journal of Oral Science (2017) 9, 104–109 OPEN www.nature.com/ijos ORIGINAL ARTICLE Identification of shared and unique gene families associated with oral clefts Noriko Funato and Masataka Nakamura Oral clefts, the most frequent congenital birth defects in humans, are multifactorial disorders caused by genetic and environmental factors. Epidemiological studies point to different etiologies underlying the oral cleft phenotypes, cleft lip (CL), CL and/or palate (CL/P) and cleft palate (CP). More than 350 genes have syndromic and/or nonsyndromic oral cleft associations in humans. Although genes related to genetic disorders associated with oral cleft phenotypes are known, a gap between detecting these associations and interpretation of their biological importance has remained. Here, using a gene ontology analysis approach, we grouped these candidate genes on the basis of different functional categories to gain insight into the genetic etiology of oral clefts. We identified different genetic profiles and found correlations between the functions of gene products and oral cleft phenotypes. Our results indicate inherent differences in the genetic etiologies that underlie oral cleft phenotypes and support epidemiological evidence that genes associated with CL/P are both developmentally and genetically different from CP only, incomplete CP, and submucous CP. The epidemiological differences among cleft phenotypes may reflect differences in the underlying genetic causes. Understanding the different causative etiologies of oral clefts is -

Review Article Association Between IRF6 Rs642961 Polymorphism and Non-Syndromic Cleft Lip with Or Without a Cleft Palate: a Systematic Review and Meta-Analysis
Int J Clin Exp Med 2018;11(4):3033-3042 www.ijcem.com /ISSN:1940-5901/IJCEM0066579 Review Article Association between IRF6 rs642961 polymorphism and non-syndromic cleft lip with or without a cleft palate: a systematic review and meta-analysis Tzu-Hui Lee1*, Tsung-Ta Liu2*, Chih-Wei Sung3,4*, Ju-Chi Ou5, Jui-Ju Yeh6, Yung-Ting Kuo1,7, Yuan-Hung Wang8,9 Departments of 1Pediatrics, 5Emergency Medicine, 6Family Medicine, 8Medical Research, Shuang Ho Hospital, Taipei Medical University, New Taipei City 23561, Taiwan; 2Graduate Institute of Medical Sciences, College of Medicine, Taipei Medical University, Taipei, Taiwan; 3Department of Emergency Medicine, National Taiwan Univer- sity Hospital Hsin-Chu Branch, Hsinchu, Taiwan; 4Department of Emergency Medicine, National Taiwan University Hospital, Taipei, Taiwan; 7Department of Pediatrics, School of Medicine, College of Medicine, Taipei Medical University, Taipei 11031, Taiwan; 9Graduate Institute of Clinical Medicine, College of Medicine, Taipei Medical University, Taipei 11031, Taiwan. *Equal contributors. Received October 2, 2017; Accepted January 29, 2018; Epub April 15, 2018; Published April 30, 2018 Abstract: Non-syndromic cleft lip with or without a cleft palate (NSCL/P) is one of the most common newborn malformations. sPrevious studies have reported that genetic variations of interferon regulatory factor 6 (IRF6) poly- morphisms are associated with NSCL/P. However, the effect sizes of individual studies still vary. Our present study is a meta-analysis to investigate the association of IRF6 rs642961 polymorphism with NSCL/P. A literature search in PubMed was performed to select eligible literatures including observational studies which evaluated association between IRF6 polymorphisms and NSCL/P. -
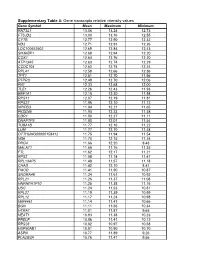
Gene Transcripts Relative Intensity Values
Supplementary Table 4: Gene transcripts relative intensity values Gene Symbol Mean Maximum Minimum RN7SL1 13.06 13.24 12.73 FTSJD2 13.00 13.16 12.55 CYTB 12.77 12.90 12.32 ND2 12.71 12.91 12.26 LOC100652902 12.69 12.84 12.43 SH3KBP1 12.68 12.94 12.20 COX1 12.64 12.76 12.20 ATP13A5 12.63 12.74 12.29 CCDC104 12.62 12.74 12.24 RPL41 12.58 12.66 12.36 TPT1 12.51 12.70 11.96 PTPRO 12.49 12.70 12.06 FN1 12.33 12.68 12.00 TLE1 12.26 12.43 11.93 EEF1A1 12.15 12.30 11.98 RPS11 12.07 12.19 11.81 RPS27 11.96 12.10 11.72 NPIPB3 11.94 12.21 11.65 FKSG49 11.94 12.23 11.58 CDR1 11.90 12.27 11.11 DNAPTP3 11.80 12.07 11.54 TUBA1B 11.77 12.16 11.22 LUM 11.77 12.10 11.48 OTTHUMG00000158412 11.75 11.94 11.54 ND6 11.70 12.15 11.24 PRG4 11.66 12.20 9.43 MALAT1 11.65 11.76 11.35 FTL 11.62 12.17 11.21 RPS2 11.58 11.74 11.47 RPL13AP5 11.48 11.57 11.18 CHAD 11.42 12.10 8.41 FMOD 11.41 11.90 10.87 SNORA48 11.34 11.61 10.92 RPL21 11.26 11.37 11.08 HNRNPA1P10 11.25 11.38 11.16 UBC 11.24 11.53 10.81 RPL27 11.19 11.39 10.99 RPL12 11.17 11.24 10.98 MIR4461 11.14 11.47 10.66 BGN 11.11 11.66 10.34 HTRA1 11.01 11.87 9.65 NEAT1 10.93 11.38 10.23 PRELP 10.86 11.41 10.12 RPS28 10.82 10.97 10.58 HSP90AB1 10.81 10.90 10.70 ASPN 10.77 11.89 9.26 PLA2G2A 10.76 11.47 9.56 H3F3A 10.69 10.86 10.41 EEF1G 10.69 10.94 10.11 C6orf48 10.64 11.02 10.35 UBA52 10.58 10.75 10.08 CTGF 10.57 11.33 9.66 MGP 10.57 11.22 10.09 YBX1 10.56 10.79 10.01 MT1X 10.56 11.89 9.88 NPC2 10.55 10.80 10.12 LAPTM4A 10.51 10.78 10.18 ITM2B 10.51 10.68 10.34 IBSP 10.50 11.50 7.17 NPIPB5 10.50 10.73 10.26 TUBA1A