Gene Transcripts Relative Intensity Values
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Browsing Genes and Genomes with Ensembl
The Bioinformatics Roadshow Tórshavn, The Faroe Islands 28-29 November 2012 BROWSING GENES AND GENOMES WITH ENSEMBL EXERCISES AND ANSWERS 1 BROWSER 3 BIOMART 8 VARIATION 13 COMPARATIVE GENOMICS 18 2 Note: These exercises are based on Ensembl version 69 (October 2012). After in future a new version has gone live, version 69 will still be available at http://e69.ensembl.org/. If your answer doesn’t correspond with the given answer, please consult the instructor. ______________________________________________________________ BROWSER ______________________________________________________________ Exercise 1 – Exploring a gene (a) Find the human F9 (coagulation factor IX) gene. On which chromosome and which strand of the genome is this gene located? How many transcripts (splice variants) have been annotated for it? (b) What is the longest transcript? How long is the protein it encodes? Has this transcript been annotated automatically (by Ensembl) or manually (by Havana)? How many exons does it have? Are any of the exons completely or partially untranslated? (c) Have a look at the external references for ENST00000218099. What is the function of F9? (d) Is it possible to monitor expression of ENST00000218099 with the ILLUMINA HumanWG_6_V2 microarray? If so, can it also be used to monitor expression of the other two transcripts? (e) In which part (i.e. the N-terminal or C-terminal half) of the protein encoded by ENST00000218099 does its peptidase activity reside? (f) Have any missense variants been discovered for the protein encoded by ENST00000218099? (g) Is there a mouse orthologue predicted for the human F9 gene? (h) If you have yourself a gene of interest, explore what information Ensembl displays about it! ______________________________________________________________ Answer (a) 8 Go to the Ensembl homepage (http://www.ensembl.org/). -

Identification of Key Pathways and Genes in Dementia Via Integrated Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.18.440371; this version posted July 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Identification of Key Pathways and Genes in Dementia via Integrated Bioinformatics Analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karnataka, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2021.04.18.440371; this version posted July 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract To provide a better understanding of dementia at the molecular level, this study aimed to identify the genes and key pathways associated with dementia by using integrated bioinformatics analysis. Based on the expression profiling by high throughput sequencing dataset GSE153960 derived from the Gene Expression Omnibus (GEO), the differentially expressed genes (DEGs) between patients with dementia and healthy controls were identified. With DEGs, we performed a series of functional enrichment analyses. Then, a protein–protein interaction (PPI) network, modules, miRNA-hub gene regulatory network and TF-hub gene regulatory network was constructed, analyzed and visualized, with which the hub genes miRNAs and TFs nodes were screened out. Finally, validation of hub genes was performed by using receiver operating characteristic curve (ROC) analysis. -

Comprehensive Analysis Reveals Novel Gene Signature in Head and Neck Squamous Cell Carcinoma: Predicting Is Associated with Poor Prognosis in Patients
5892 Original Article Comprehensive analysis reveals novel gene signature in head and neck squamous cell carcinoma: predicting is associated with poor prognosis in patients Yixin Sun1,2#, Quan Zhang1,2#, Lanlin Yao2#, Shuai Wang3, Zhiming Zhang1,2 1Department of Breast Surgery, The First Affiliated Hospital of Xiamen University, School of Medicine, Xiamen University, Xiamen, China; 2School of Medicine, Xiamen University, Xiamen, China; 3State Key Laboratory of Cellular Stress Biology, School of Life Sciences, Xiamen University, Xiamen, China Contributions: (I) Conception and design: Y Sun, Q Zhang; (II) Administrative support: Z Zhang; (III) Provision of study materials or patients: Y Sun, Q Zhang; (IV) Collection and assembly of data: Y Sun, L Yao; (V) Data analysis and interpretation: Y Sun, S Wang; (VI) Manuscript writing: All authors; (VII) Final approval of manuscript: All authors. #These authors contributed equally to this work. Correspondence to: Zhiming Zhang. Department of Surgery, The First Affiliated Hospital of Xiamen University, Xiamen, China. Email: [email protected]. Background: Head and neck squamous cell carcinoma (HNSC) remains an important public health problem, with classic risk factors being smoking and excessive alcohol consumption and usually has a poor prognosis. Therefore, it is important to explore the underlying mechanisms of tumorigenesis and screen the genes and pathways identified from such studies and their role in pathogenesis. The purpose of this study was to identify genes or signal pathways associated with the development of HNSC. Methods: In this study, we downloaded gene expression profiles of GSE53819 from the Gene Expression Omnibus (GEO) database, including 18 HNSC tissues and 18 normal tissues. -

This Item Is the Archived Peer-Reviewed Author-Version Of
This item is the archived peer-reviewed author-version of: Deiodinase knockdown affects zebrafish eye development at the level of gene expression, morphology and function Reference: Houbrechts Anne M., Vergauwen Lucia, Bagci Enise, Van houcke Jolien, Heijlen Marjolein, Kulemeka Bernard, Hyde David R., Knapen Dries, Darras Veerle M..- Deiodinase knockdown affects zebrafish eye development at the level of gene expression, morphology and function Molecular and cellular endocrinology - ISSN 0303-7207 - 424(2016), p. 81-93 Full text (Publishers DOI): http://dx.doi.org/doi:10.1016/j.mce.2016.01.018 To cite this reference: http://hdl.handle.net/10067/1314550151162165141 Institutional repository IRUA 1 Deiodinase knockdown affects zebrafish eye development at the level 2 of gene expression, morphology and function. 3 Anne M. Houbrechts1*, Lucia Vergauwen2*, Enise Bagci2,3, Jolien Van houcke1, Marjolein 4 Heijlen1, Bernard Kulemeka4, David R. Hyde4, Dries Knapen2, Veerle M. Darras1 5 1 Laboratory of Comparative Endocrinology, Department of Biology, Division of 6 Animal Physiology and Neurobiology, KU Leuven, 3000 Leuven, Belgium 7 2 Zebrafishlab, Veterinary Physiology and Biochemistry, Department of Veterinary 8 Sciences, University of Antwerp, Universiteitsplein 1, 2610 Wilrijk, Belgium 9 3 Systemic Physiological & Ecotoxicological Research, Department of Biology, 10 University of Antwerp, Universiteitsplein 1, 2610 Wilrijk, Belgium 11 4 Department of Biological Sciences and Center for Zebrafish Research, University of 12 Notre Dame, Notre Dame, -

Original Article LIN28B Suppresses Microrna Let-7B Expression to Promote CD44+/LIN28B+ Human Pancreatic Cancer Stem Cell Proliferation and Invasion
Am J Cancer Res 2015;5(9):2643-2659 www.ajcr.us /ISSN:2156-6976/ajcr0008500 Original Article LIN28B suppresses microRNA let-7b expression to promote CD44+/LIN28B+ human pancreatic cancer stem cell proliferation and invasion Yebo Shao1*, Lei Zhang1*, Lei Cui2*, Wenhui Lou1, Dansong Wang1, Weiqi Lu1, Dayong Jin1, Te Liu3 1Department of General Surgery, Zhongshan Hospital, Fudan University, Shanghai 200032, China; 2Department of General Surgery, Jiangsu University Affiliated Hospital, Zhengjiang 212000, China; 3Shanghai Geriatric Institute of Chinese Medicine, Longhua Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai 200031, China. *Equal contributors. Received March 27, 2015; Accepted July 5, 2015; Epub August 15, 2015; Published September 1, 2015 Abstract: Although the highly proliferative, migratory, and multi-drug resistant phenotype of human pancreatic can- cer stem cells (PCSCs) is well characterized, knowledge of their biological mechanisms is limited. We used CD44 and LIN28B as markers to screen, isolate, and enrich CSCs from human primary pancreatic cancer. Using flow cytometry, we identified a human primary pancreatic cancer cell (PCC) subpopulation expressing high levels of both CD44 and LIN28B. CD44+/LIN28B+ PCSCs expressed high levels of stemness marker genes and possessed higher migratory and invasive ability than CD44-/LIN28B- PCCs. CD44+/LIN28B+ PCSCs were more resistant to growth inhibition induced by the chemotherapeutic drugs cisplatin and gemcitabine hydrochloride, and readily established tumors in vivo in a relatively short time. Moreover, microarray analysis revealed significant differences between the cDNA expression patterns of CD44+/LIN28B+ PCSCs and CD44-/LIN28B- PCCs. Following siRNA interference of endogenous LIN28B gene expression in CD44+/LIN28B+ PCSCs, not only was their proliferation decreased, there was also cell cycle arrest due to suppression of cyclin D1 expression following the stimulation of miRNA let-7b expression. -

A Brain-Derived Neurotrophic Factor
www.nature.com/scientificreports OPEN A Brain-Derived Neurotrophic Factor Mimetic Is Sufcient to Restore Cone Photoreceptor Visual Received: 17 January 2017 Accepted: 25 August 2017 Function in an Inherited Blindness Published: xx xx xxxx Model Conor Daly1, Lisa Shine1, Theresa Hefernan1, Sudhakar Deeti1, Alison L. Reynolds 1, John J. O’Connor1, Eugène T. Dillon 1,2, David J. Dufy 3,5, Walter Kolch 1,3,4, Gerard Cagney1 & Breandán N. Kennedy 1 Controversially, histone deacetylase inhibitors (HDACi) are in clinical trial for the treatment of inherited retinal degeneration. Utilizing the zebrafsh dyeucd6 model, we determined if treatment with HDACi can rescue cone photoreceptor-mediated visual function. dye exhibit defective visual behaviour and retinal morphology including ciliary marginal zone (CMZ) cell death and decreased photoreceptor outer segment (OS) length, as well as gross morphological defects including hypopigmentation and pericardial oedema. HDACi treatment of dye results in signifcantly improved optokinetic (OKR) (~43 fold, p < 0.001) and visualmotor (VMR) (~3 fold, p < 0.05) responses. HDACi treatment rescued gross morphological defects and reduced CMZ cell death by 80%. Proteomic analysis of dye eye extracts suggested BDNF-TrkB and Akt signaling as mediators of HDACi rescue in our dataset. Co- treatment with the TrkB antagonist ANA-12 blocked HDACi rescue of visual function and associated Akt phosphorylation. Notably, sole treatment with a BDNF mimetic, 7,8-dihydroxyfavone hydrate, signifcantly rescued dye visual function (~58 fold increase in OKR, p < 0.001, ~3 fold increase in VMR, p < 0.05). In summary, HDACi and a BDNF mimetic are sufcient to rescue retinal cell death and visual function in a vertebrate model of inherited blindness. -
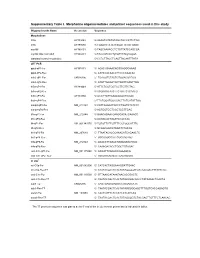
Supplementary Table I. Morpholino Oligonucleotides and Primer Sequences Used in This Study
Supplementary Table I. Morpholino oligonucleotides and primer sequences used in this study Oligonucleotide Name Accession Sequence Morpholinos tlr5a AY389449 5'-AAAGTGTATGTAGCTGCCATTCTGG tlr5b AY389450 5'-TGAATGTATATCCCATTCTGTGAGC myd88 AY388401 5'-TAGCAAAACCTCTGTTATCCAGCGA myd88 5bp mismatch AY388401 5'-TAcCAtAACCTgTGTTATCgAGgGA standard control morpholino 5'-CCTCTTACCTCAGTTACAATTTATA qRT-PCR ppial-qP1-Fw AY391451 5’- ACACTGAAACACGGAGGCAAAG ppial-qP2-Rev 5’- CATCCACAACCTTCCCGAACAC irak3-qP1-Fw CK026195 5’- TGAGGTCTACTGTGGACGATGG irak3-qP2-Rev 5’- ATGTTAGGATGCTGGTTGAGTTGG tlr5a-qP1-Fw AY389449 5’-ATTCTGGTGGTGCTTGTTGTAG tlr5a-qP2-Rev 5’-ACGAGGTAACTTCTGTTCTCAATG tlr5b-qP3-Fw AY389450 5’-GCGTTGTTGAAGAGGCTGGAC tlr5b-qP4-Rev 5’-TTCTGGATGGCCACTTCTCATATTGG mmp9-qP3-Fw NM_213123 5’-CATTAAAGATGCCCTGATGTATCCC mmp9-qP4-Rev 5’-AGTGGTGGTCCGTGGTTGAG il1b-qP1-Fw NM_212844 5’-GAACAGAATGAAGCACATCAAACC il1b-qP2-Rev 5’-ACGGCACTGAATCCACCAC il8-qP1-Fw XM_001342570 5’-TGTGTTATTGTTTTCCTGGCATTTC il8-qP2-Rev 5’-GCGACAGCGTGGATCTACAG ifn1-qP3-Fw NM_207640 5’- TTAATACACGCAAAGATGAGAACTC ifn1-qP4-Rev 5’- GCCAAGCCATTCGCAAGTAG tnfa-qP5-Fw NM_212829 5’- AGACCTTAGACTGGAGAGATGAC tnfa-qP6-Rev 5’- CAAAGACACCTGGCTGTAGAC cxcl-C1c-qP1-Fw NM_001115060 5’- GGCATTCACACCCAAAGCG cxcl-C1c-qP2_Rev 5’- GCGAGCACGATTCACGAGAG * In situ ccl-C5a-Fw NM_001082906 5’- CATCACTAGGAAAGGATTGAAC ccl-C5a-Rev-T7 5’- TAATACGACTCACTATAGGGGATGTCAAAGACTTTATTCAC cxcl-C1c-Fw NM_001115060 5’- GTTAAACATAAATAACACCGACTC cxcl-C1c-Rev-T7 5’- TAATACGACTCACTATAGGGACACCCTATAAAACTGAGTA irak3-Fw CK026195 5’- CAGTGAGAGAGGCATGAAACATC -

SUPPLEMENTARY DATA Retinoic Acid Mediates Visceral-Specific
SUPPLEMENTARY DATA Retinoic Acid Mediates Visceral-specific Adipogenic Defects of Human Adipose-derived Stem Cells Kosuke Takeda, Sandhya Sriram, Xin Hui Derryn Chan, Wee Kiat Ong, Chia Rou Yeo, Betty Tan, Seung-Ah Lee, Kien Voon Kong, Shawn Hoon, Hongfeng Jiang, Jason J. Yuen, Jayakumar Perumal, Madhur Agrawal, Candida Vaz, Jimmy So, Asim Shabbir, William S. Blaner, Malini Olivo, Weiping Han, Vivek Tanavde, Sue-Anne Toh, and Shigeki Sugii Supplementary Experimental Procedures RNA-Seq experiment and analysis Total RNA was extracted from SC and VS fat depots using Qiagen RNeasy plus kit. Poly-A mRNA was then enriched from ~5 µg of total RNA with oligo dT beads (Life Technologies). Approximately 100 ng of poly-A mRNA recovered was used to construct multiplexed strand-specific RNA-seq libraries as per manufacturer’s instruction (NEXTflexTM Rapid Directional RNA-SEQ Kit (dUTP-Based) v2). Individual library quality was assessed with an Agilent 2100 Bioanalyzer and quantified with a QuBit 2.0 fluorometer before pooling for sequencing on Illumina HiSeq 2000 (1x101 bp read). The pooled libraries were quantified using the KAPA quantification kit (KAPA Biosystems) prior to cluster formation. Fastq formatted reads were processed with Trimmomatic to remove adapter sequences and trim low quality bases (LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36). Reads were aligned to the human genome (hg19) using Tophat version 2 (settings --no-coverage-search --library- type=fr-firststrand). Feature read counts were generated using htseq-count (Python package HTSeq default union-counting mode, strand=reverse). Differential Expression analysis was performed using the edgeR package in both ‘classic’ and generalized linear model (glm) modes to contrast SC and VS adipose tissues from 8 non-diabetic patients. -

C21orf62 CRISPR/Cas9 KO Plasmid (H): Sc-406451
SANTA CRUZ BIOTECHNOLOGY, INC. C21orf62 CRISPR/Cas9 KO Plasmid (h): sc-406451 BACKGROUND APPLICATIONS The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and C21orf62 CRISPR/Cas9 KO Plasmid (h) is recommended for the disruption of CRISPR-associated protein (Cas9) system is an adaptive immune response gene expression in human cells. defense mechanism used by archea and bacteria for the degradation of foreign genetic material (4,6). This mechanism can be repurposed for other 20 nt non-coding RNA sequence: guides Cas9 functions, including genomic engineering for mammalian systems, such as to a specific target location in the genomic DNA gene knockout (KO) (1,2,3,5). CRISPR/Cas9 KO Plasmid products enable the U6 promoter: drives gRNA scaffold: helps Cas9 identification and cleavage of specific genes by utilizing guide RNA (gRNA) expression of gRNA bind to target DNA sequences derived from the Genome-scale CRISPR Knock-Out (GeCKO) v2 library developed in the Zhang Laboratory at the Broad Institute (3,5). Termination signal Green Fluorescent Protein: to visually REFERENCES verify transfection CRISPR/Cas9 Knockout Plasmid CBh (chicken β-Actin 1. Cong, L., et al. 2013. Multiplex genome engineering using CRISPR/Cas hybrid) promoter: drives systems. Science 339: 819-823. 2A peptide: expression of Cas9 allows production of both Cas9 and GFP from the 2. Mali, P., et al. 2013. RNA-guided human genome engineering via Cas9. same CBh promoter Science 339: 823-826. Nuclear localization signal 3. Ran, F.A., et al. 2013. Genome enginee ring using the CRISPR-Cas9 system. Nuclear localization signal SpCas9 ribonuclease Nat. Protoc. 8: 2281-2308. -

Supplementary Table S2. List of Genes with Expression That Is Positively Correlated (Pearson Correlation Coefficient P>0.3)
Supplementary Table S2. List of genes with expression that is positively correlated (Pearson correlation coefficient p>0.3) with HOXA9 expression in the study by Sun et al (1). Most HOXA genes (indicated by bold face) show highly significant positive correlation with HOXA9 expression, except for HOXA6 and HOXA13, similar to our findings in the UCSF and MDA tumor sets. Note that HOXA11, which did not demonstrate a statistically significant positive correlation with HOXA9 expression in both the UCSF and MDA tumor sets, demonstrates a substantially lower correlation coefficient relative to the other HOXA genes that positively correlate with HOXA9 expression in the study by Sun et al. These data were obtained from the online ONCOMINE database (www.oncomine.org) (2) searching for transcripts positively correlated with HOXA9 expression. Gene Name Gene Symbol Reporter ID Correlation (p) homeobox A9 HOXA9 214651_s_at .7682 homeobox A9 HOXA9 209905_at .7682 homeobox A10 HOXA10 213150_at .6058 homeobox A7 HOXA7 235753_at .6058 homeobox A10 HOXA10 213147_at .6058 homeobox A7 HOXA7 206847_s_at .6058 homeobox A4 HOXA4 206289_at .5741 homeobox A2 HOXA2 1557051_s_at .5379 homeobox A1 HOXA1 214639_s_at .5379 homeobox A3 HOXA3 235521_at .5379 homeobox B2 HOXB2 205453_at .5379 homeobox B3 HOXB3 228904_at .5379 EST EST 1555907_at .5379 homeobox A4 HOXA4 230080_at .5379 homeobox A2 HOXA2 228642_at .5379 homeobox A2 HOXA2 1557050_at .5379 homeobox A5 HOXA5 213844_at .5379 homeobox A2 HOXA2 214457_at .5113 homeobox B7 HOXB7 204778_x_at .5042 homeobox C6 HOXC6 -

SUPPLEMENTARY MATERIALS and METHODS PBMC Transcriptomics
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut SUPPLEMENTARY MATERIALS AND METHODS PBMC transcriptomics identifies immune-metabolism disorder during the development of HBV-ACLF Contents l Supplementary methods l Supplementary Figure 1 l Supplementary Figure 2 l Supplementary Figure 3 l Supplementary Figure 4 l Supplementary Figure 5 l Supplementary Table 1 l Supplementary Table 2 l Supplementary Table 3 l Supplementary Table 4 l Supplementary Tables 5-14 l Supplementary Table 15 l Supplementary Table 16 l Supplementary Table 17 Li J, et al. Gut 2021;0:1–13. doi: 10.1136/gutjnl-2020-323395 BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut SUPPLEMENTARY METHODS Test for HBV DNA The levels of HBV DNA were detected using real-time PCR with a COBAS® AmpliPrep/COBAS® TaqMan 48 System (Roche, Basel, Switzerland) and HBV Test v2.0. Criteria for diagnosing cirrhosis Pathology The gold standard for the diagnosis of cirrhosis is a liver biopsy obtained through a percutaneous or transjugular approach.1 Ultrasonography was performed 2-4 hours before biopsy. Liver biopsy specimens were obtained by experienced physicians. Percutaneous transthoracic puncture of the liver was performed according to the standard criteria. After biopsy, patients were monitored in the hospital with periodic analyses of haematocrit and other vital signs for 24 hours. Cirrhosis was diagnosed according to the globally agreed upon criteria.2 Cirrhosis is defined based on its pathological features under a microscope: (a) the presence of parenchymal nodules, (b) differences in liver cell size and appearance, (c) fragmentation of the biopsy specimen, (d) fibrous septa, and (d) an altered architecture and vascular relationships.