An Extended Family of Novel Vertebrate Photopigments Is Widely Expressed and Displays a Diversity of Function
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Browsing Genes and Genomes with Ensembl
The Bioinformatics Roadshow Tórshavn, The Faroe Islands 28-29 November 2012 BROWSING GENES AND GENOMES WITH ENSEMBL EXERCISES AND ANSWERS 1 BROWSER 3 BIOMART 8 VARIATION 13 COMPARATIVE GENOMICS 18 2 Note: These exercises are based on Ensembl version 69 (October 2012). After in future a new version has gone live, version 69 will still be available at http://e69.ensembl.org/. If your answer doesn’t correspond with the given answer, please consult the instructor. ______________________________________________________________ BROWSER ______________________________________________________________ Exercise 1 – Exploring a gene (a) Find the human F9 (coagulation factor IX) gene. On which chromosome and which strand of the genome is this gene located? How many transcripts (splice variants) have been annotated for it? (b) What is the longest transcript? How long is the protein it encodes? Has this transcript been annotated automatically (by Ensembl) or manually (by Havana)? How many exons does it have? Are any of the exons completely or partially untranslated? (c) Have a look at the external references for ENST00000218099. What is the function of F9? (d) Is it possible to monitor expression of ENST00000218099 with the ILLUMINA HumanWG_6_V2 microarray? If so, can it also be used to monitor expression of the other two transcripts? (e) In which part (i.e. the N-terminal or C-terminal half) of the protein encoded by ENST00000218099 does its peptidase activity reside? (f) Have any missense variants been discovered for the protein encoded by ENST00000218099? (g) Is there a mouse orthologue predicted for the human F9 gene? (h) If you have yourself a gene of interest, explore what information Ensembl displays about it! ______________________________________________________________ Answer (a) 8 Go to the Ensembl homepage (http://www.ensembl.org/). -

G Protein-Coupled Receptors
S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. British Journal of Pharmacology (2015) 172, 5744–5869 THE CONCISE GUIDE TO PHARMACOLOGY 2015/16: G protein-coupled receptors Stephen PH Alexander1, Anthony P Davenport2, Eamonn Kelly3, Neil Marrion3, John A Peters4, Helen E Benson5, Elena Faccenda5, Adam J Pawson5, Joanna L Sharman5, Christopher Southan5, Jamie A Davies5 and CGTP Collaborators 1School of Biomedical Sciences, University of Nottingham Medical School, Nottingham, NG7 2UH, UK, 2Clinical Pharmacology Unit, University of Cambridge, Cambridge, CB2 0QQ, UK, 3School of Physiology and Pharmacology, University of Bristol, Bristol, BS8 1TD, UK, 4Neuroscience Division, Medical Education Institute, Ninewells Hospital and Medical School, University of Dundee, Dundee, DD1 9SY, UK, 5Centre for Integrative Physiology, University of Edinburgh, Edinburgh, EH8 9XD, UK Abstract The Concise Guide to PHARMACOLOGY 2015/16 provides concise overviews of the key properties of over 1750 human drug targets with their pharmacology, plus links to an open access knowledgebase of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties. The full contents can be found at http://onlinelibrary.wiley.com/doi/ 10.1111/bph.13348/full. G protein-coupled receptors are one of the eight major pharmacological targets into which the Guide is divided, with the others being: ligand-gated ion channels, voltage-gated ion channels, other ion channels, nuclear hormone receptors, catalytic receptors, enzymes and transporters. These are presented with nomenclature guidance and summary information on the best available pharmacological tools, alongside key references and suggestions for further reading. -

Multi-Functionality of Proteins Involved in GPCR and G Protein Signaling: Making Sense of Structure–Function Continuum with In
Cellular and Molecular Life Sciences (2019) 76:4461–4492 https://doi.org/10.1007/s00018-019-03276-1 Cellular andMolecular Life Sciences REVIEW Multi‑functionality of proteins involved in GPCR and G protein signaling: making sense of structure–function continuum with intrinsic disorder‑based proteoforms Alexander V. Fonin1 · April L. Darling2 · Irina M. Kuznetsova1 · Konstantin K. Turoverov1,3 · Vladimir N. Uversky2,4 Received: 5 August 2019 / Revised: 5 August 2019 / Accepted: 12 August 2019 / Published online: 19 August 2019 © Springer Nature Switzerland AG 2019 Abstract GPCR–G protein signaling system recognizes a multitude of extracellular ligands and triggers a variety of intracellular signal- ing cascades in response. In humans, this system includes more than 800 various GPCRs and a large set of heterotrimeric G proteins. Complexity of this system goes far beyond a multitude of pair-wise ligand–GPCR and GPCR–G protein interactions. In fact, one GPCR can recognize more than one extracellular signal and interact with more than one G protein. Furthermore, one ligand can activate more than one GPCR, and multiple GPCRs can couple to the same G protein. This defnes an intricate multifunctionality of this important signaling system. Here, we show that the multifunctionality of GPCR–G protein system represents an illustrative example of the protein structure–function continuum, where structures of the involved proteins represent a complex mosaic of diferently folded regions (foldons, non-foldons, unfoldons, semi-foldons, and inducible foldons). The functionality of resulting highly dynamic conformational ensembles is fne-tuned by various post-translational modifcations and alternative splicing, and such ensembles can undergo dramatic changes at interaction with their specifc partners. -

Journal 37.Pdf
Biomaterials 34 (2013) 1911e1920 Contents lists available at SciVerse ScienceDirect Biomaterials journal homepage: www.elsevier.com/locate/biomaterials The activation of directional stem cell motility by green light-emitting diode irradiation Wei-Kee Ong a,1, How-Foo Chen b,1, Cheng-Ting Tsai b, Yun-Ju Fu a, Yi-Shan Wong a, Da-Jen Yen c, Tzu-Hao Chang d,e, Hsien-Da Huang d, Oscar Kuang-Sheng Lee f, Shu Chien g, Jennifer Hui-Chun Ho a,h,i,* a Center for Stem Cell Research, Wan Fang Hospital, Taipei Medical University, Taipei, Taiwan b Institute of Biophotonics, National Yang-Ming University, Taipei, Taiwan c Department of Material Science and Engineering, National Tsing Hua University, HsinChu, Taiwan d Institute of Bioinformatics and Systems Biology, National Chiao Tung University, HsinChu, Taiwan e Graduate Institute of Biomedical Informatics, Taipei Medical University, Taiwan f Institute of Clinical Medicine, National Yang-Ming University, Taiwan g Departments of Bioengineering and Medicine, Institute of Engineering in Medicine, UC San Diego, La Jolla, CA, USA h Graduate Institute of Clinical Medicine, Taipei Medical University, Taipei, Taiwan i Department of Ophthalmology, Wan Fang Hospital, Taipei Medical University, Taipei, Taiwan article info abstract Article history: Light-emitting diode (LED) irradiation is potentially a photostimulator to manipulate cell behavior by Received 12 November 2012 opsin-triggered phototransduction and thermal energy supply in living cells. Directional stem cell Accepted 29 November 2012 motility is critical for the efficiency and specificity of stem cells in tissue repair. We explored that green Available online 19 December 2012 LED (530 nm) irradiation directed the human orbital fat stem cells (OFSCs) to migrate away from the LED light source through activation of extracellular signal-regulated kinases (ERK)/MAP kinase/p38 signaling Keywords: pathway. -

Evolutionary History of Teleost Intron-Containing and Intron-Less
www.nature.com/scientificreports OPEN Evolutionary history of teleost intron-containing and intron-less rhodopsin genes Received: 15 February 2019 Chihiro Fujiyabu1, Keita Sato 2, Ni Made Laksmi Utari2,3, Hideyo Ohuchi2, Accepted: 9 July 2019 Yoshinori Shichida1,4,5 & Takahiro Yamashita1,5 Published: xx xx xxxx Recent progress in whole genome sequencing has revealed that animals have various kinds of opsin genes for photoreception. Among them, most opsin genes have introns in their coding regions. However, it has been known for a long time that teleost retinas express intron-less rhodopsin genes, which are presumed to have been formed by retroduplication from an ancestral intron-containing rhodopsin gene. In addition, teleosts have an intron-containing rhodopsin gene (exo-rhodopsin) exclusively for pineal photoreception. In this study, to unravel the evolutionary origin of the two teleost rhodopsin genes, we analyzed the rhodopsin genes of non-teleost fshes in the Actinopterygii. The phylogenetic analysis of full-length sequences of bichir, sturgeon and gar rhodopsins revealed that retroduplication of the rhodopsin gene occurred after branching of the bichir lineage. In addition, analysis of the tissue distribution and the molecular properties of bichir, sturgeon and gar rhodopsins showed that the abundant and exclusive expression of intron-containing rhodopsin in the pineal gland and the short lifetime of its meta II intermediate, which leads to optimization for pineal photoreception, were achieved after branching of the gar lineage. Based on these results, we propose a stepwise evolutionary model of teleost intron-containing and intron-less rhodopsin genes. Opsins are photoreceptive molecules that universally underlie the molecular basis of visual and non-visual pho- toreception in animals1–3. -

The Vertebrate Ancestral Repertoire of Visual Opsins, Transducin Alpha Subunits and Oxytocin/Vasopressin Receptors Was Establish
Lagman et al. BMC Evolutionary Biology 2013, 13:238 http://www.biomedcentral.com/1471-2148/13/238 RESEARCH ARTICLE Open Access The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/ vasopressin receptors was established by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications David Lagman1†, Daniel Ocampo Daza1†, Jenny Widmark1,XesúsMAbalo1, Görel Sundström1,2 and Dan Larhammar1* Abstract Background: Vertebrate color vision is dependent on four major color opsin subtypes: RH2 (green opsin), SWS1 (ultraviolet opsin), SWS2 (blue opsin), and LWS (red opsin). Together with the dim-light receptor rhodopsin (RH1), these form the family of vertebrate visual opsins. Vertebrate genomes contain many multi-membered gene families that can largely be explained by the two rounds of whole genome duplication (WGD) in the vertebrate ancestor (2R) followed by a third round in the teleost ancestor (3R). Related chromosome regions resulting from WGD or block duplications are said to form a paralogon. We describe here a paralogon containing the genes for visual opsins, the G-protein alpha subunit families for transducin (GNAT) and adenylyl cyclase inhibition (GNAI), the oxytocin and vasopressin receptors (OT/VP-R), and the L-type voltage-gated calcium channels (CACNA1-L). Results: Sequence-based phylogenies and analyses of conserved synteny show that the above-mentioned gene families, and many neighboring gene families, expanded in the early vertebrate WGDs. This allows us to deduce the following evolutionary scenario: The vertebrate ancestor had a chromosome containing the genes for two visual opsins, one GNAT, one GNAI, two OT/VP-Rs and one CACNA1-L gene. -

Identification of Estradiol/Era-Regulated Genes in the Mouse Pituitary
309 Identification of estradiol/ERa-regulated genes in the mouse pituitary Hyun Joon Kim1,2,*, Mary C Gieske1,3,*, Kourtney L Trudgen1,*, Susan Hudgins-Spivey1, Beob Gyun Kim4, Andree Krust5,6, Pierre Chambon5,6, Jae-Wook Jeong7, Eric Blalock8 and CheMyong Ko1,3 1Division of Reproductive Sciences, Department of Clinical Sciences, University of Kentucky, Lexington, Kentucky 40536, USA 2Department of Anatomy and Neurobiology, Institute of Health Sciences, Medical Research Center for Neural Dysfunction, School of Medicine, Gyeongsang National University, Jinju, Korea 3Department of Biology, University of Kentucky, Lexington, Kentucky 40536, USA 4Department of Animal Science and Environment, Konkuk University, Seoul 143-701, Korea 5Institut de Genetique et de Biologie Moleculaire et Cellulaire (CNRS, INSERM, ULP, College de France), 67404 Illkirch Cedex, Strasbourg, France 6Institut Clinique de la Souris, BP10142, 67404 Illkirch Cedex, Strasbourg, France 7Department of Obstetrics, Gynecology and Reproductive Biology, Michigan State University College of Human Medicine, Grand Rapids, Michigan 49503, USA 8Department of Molecular and Biomedical Pharmacology, University of Kentucky, Lexington, Kentucky 40536, USA (Correspondence should be addressed to C Ko who is now at Division of Clinical and Reproductive Sciences, College of Health Sciences, University of Kentucky, Lexington, Kentucky 40536, USA; Email: [email protected]) *(H J Kim, M C Gieske, and K L Trudgen contributed equally to this work) Abstract Estrogen acts to prime the pituitary prior to the GnRH- pathway in the pituitary. This approach substantiates ERa induced LH surge by undiscovered mechanisms. This study regulation of membrane potential regulators and intracellular aimed to identify the key components that mediate estrogen vesicle transporters, among others, but not the basic action in priming the pituitary. -
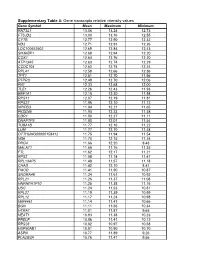
Gene Transcripts Relative Intensity Values
Supplementary Table 4: Gene transcripts relative intensity values Gene Symbol Mean Maximum Minimum RN7SL1 13.06 13.24 12.73 FTSJD2 13.00 13.16 12.55 CYTB 12.77 12.90 12.32 ND2 12.71 12.91 12.26 LOC100652902 12.69 12.84 12.43 SH3KBP1 12.68 12.94 12.20 COX1 12.64 12.76 12.20 ATP13A5 12.63 12.74 12.29 CCDC104 12.62 12.74 12.24 RPL41 12.58 12.66 12.36 TPT1 12.51 12.70 11.96 PTPRO 12.49 12.70 12.06 FN1 12.33 12.68 12.00 TLE1 12.26 12.43 11.93 EEF1A1 12.15 12.30 11.98 RPS11 12.07 12.19 11.81 RPS27 11.96 12.10 11.72 NPIPB3 11.94 12.21 11.65 FKSG49 11.94 12.23 11.58 CDR1 11.90 12.27 11.11 DNAPTP3 11.80 12.07 11.54 TUBA1B 11.77 12.16 11.22 LUM 11.77 12.10 11.48 OTTHUMG00000158412 11.75 11.94 11.54 ND6 11.70 12.15 11.24 PRG4 11.66 12.20 9.43 MALAT1 11.65 11.76 11.35 FTL 11.62 12.17 11.21 RPS2 11.58 11.74 11.47 RPL13AP5 11.48 11.57 11.18 CHAD 11.42 12.10 8.41 FMOD 11.41 11.90 10.87 SNORA48 11.34 11.61 10.92 RPL21 11.26 11.37 11.08 HNRNPA1P10 11.25 11.38 11.16 UBC 11.24 11.53 10.81 RPL27 11.19 11.39 10.99 RPL12 11.17 11.24 10.98 MIR4461 11.14 11.47 10.66 BGN 11.11 11.66 10.34 HTRA1 11.01 11.87 9.65 NEAT1 10.93 11.38 10.23 PRELP 10.86 11.41 10.12 RPS28 10.82 10.97 10.58 HSP90AB1 10.81 10.90 10.70 ASPN 10.77 11.89 9.26 PLA2G2A 10.76 11.47 9.56 H3F3A 10.69 10.86 10.41 EEF1G 10.69 10.94 10.11 C6orf48 10.64 11.02 10.35 UBA52 10.58 10.75 10.08 CTGF 10.57 11.33 9.66 MGP 10.57 11.22 10.09 YBX1 10.56 10.79 10.01 MT1X 10.56 11.89 9.88 NPC2 10.55 10.80 10.12 LAPTM4A 10.51 10.78 10.18 ITM2B 10.51 10.68 10.34 IBSP 10.50 11.50 7.17 NPIPB5 10.50 10.73 10.26 TUBA1A -

Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Amy Sue Bogard University of Tennessee Health Science Center
University of Tennessee Health Science Center UTHSC Digital Commons Theses and Dissertations (ETD) College of Graduate Health Sciences 12-2013 Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Amy Sue Bogard University of Tennessee Health Science Center Follow this and additional works at: https://dc.uthsc.edu/dissertations Part of the Medical Cell Biology Commons, and the Medical Molecular Biology Commons Recommended Citation Bogard, Amy Sue , "Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells" (2013). Theses and Dissertations (ETD). Paper 330. http://dx.doi.org/10.21007/etd.cghs.2013.0029. This Dissertation is brought to you for free and open access by the College of Graduate Health Sciences at UTHSC Digital Commons. It has been accepted for inclusion in Theses and Dissertations (ETD) by an authorized administrator of UTHSC Digital Commons. For more information, please contact [email protected]. Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells Document Type Dissertation Degree Name Doctor of Philosophy (PhD) Program Biomedical Sciences Track Molecular Therapeutics and Cell Signaling Research Advisor Rennolds Ostrom, Ph.D. Committee Elizabeth Fitzpatrick, Ph.D. Edwards Park, Ph.D. Steven Tavalin, Ph.D. Christopher Waters, Ph.D. DOI 10.21007/etd.cghs.2013.0029 Comments Six month embargo expired June 2014 This dissertation is available at UTHSC Digital Commons: https://dc.uthsc.edu/dissertations/330 Adenylyl Cyclase 2 Selectively Regulates IL-6 Expression in Human Bronchial Smooth Muscle Cells A Dissertation Presented for The Graduate Studies Council The University of Tennessee Health Science Center In Partial Fulfillment Of the Requirements for the Degree Doctor of Philosophy From The University of Tennessee By Amy Sue Bogard December 2013 Copyright © 2013 by Amy Sue Bogard. -

(Raav)-Vector Elements in Ocular Gene Therapy Clinical Trials and Transgene Expression and Bioactivity Assays
International Journal of Molecular Sciences Review Recombinant Adeno-Associated Viral Vectors (rAAV)-Vector Elements in Ocular Gene Therapy Clinical Trials and Transgene Expression and Bioactivity Assays Thilo M. Buck 1 and Jan Wijnholds 1,2,* 1 Department of Ophthalmology, Leiden University Medical Center (LUMC), 2333 ZC Leiden, The Netherlands; [email protected] 2 Netherlands Institute of Neuroscience, Royal Netherlands Academy of Arts and Sciences (KNAW), 1105 BA Amsterdam, The Netherlands * Correspondence: [email protected]; Tel.: +31-71-52-69269 Received: 20 May 2020; Accepted: 10 June 2020; Published: 12 June 2020 Abstract: Inherited retinal dystrophies and optic neuropathies cause chronic disabling loss of visual function. The development of recombinant adeno-associated viral vectors (rAAV) gene therapies in all disease fields have been promising, but the translation to the clinic has been slow. The safety and efficacy profiles of rAAV are linked to the dose of applied vectors. DNA changes in the rAAV gene cassette affect potency, the expression pattern (cell-specificity), and the production yield. Here, we present a library of rAAV vectors and elements that provide a workflow to design novel vectors. We first performed a meta-analysis on recombinant rAAV elements in clinical trials (2007–2020) for ocular gene therapies. We analyzed 33 unique rAAV gene cassettes used in 57 ocular clinical trials. The rAAV gene therapy vectors used six unique capsid variants, 16 different promoters, and six unique polyadenylation sequences. Further, we compiled a list of promoters, enhancers, and other sequences used in current rAAV gene cassettes in preclinical studies. Then, we give an update on pro-viral plasmid backbones used to produce the gene therapy vectors, inverted terminal repeats, production yield, and rAAV safety considerations. -

This Item Is the Archived Peer-Reviewed Author-Version Of
This item is the archived peer-reviewed author-version of: Deiodinase knockdown affects zebrafish eye development at the level of gene expression, morphology and function Reference: Houbrechts Anne M., Vergauwen Lucia, Bagci Enise, Van houcke Jolien, Heijlen Marjolein, Kulemeka Bernard, Hyde David R., Knapen Dries, Darras Veerle M..- Deiodinase knockdown affects zebrafish eye development at the level of gene expression, morphology and function Molecular and cellular endocrinology - ISSN 0303-7207 - 424(2016), p. 81-93 Full text (Publishers DOI): http://dx.doi.org/doi:10.1016/j.mce.2016.01.018 To cite this reference: http://hdl.handle.net/10067/1314550151162165141 Institutional repository IRUA 1 Deiodinase knockdown affects zebrafish eye development at the level 2 of gene expression, morphology and function. 3 Anne M. Houbrechts1*, Lucia Vergauwen2*, Enise Bagci2,3, Jolien Van houcke1, Marjolein 4 Heijlen1, Bernard Kulemeka4, David R. Hyde4, Dries Knapen2, Veerle M. Darras1 5 1 Laboratory of Comparative Endocrinology, Department of Biology, Division of 6 Animal Physiology and Neurobiology, KU Leuven, 3000 Leuven, Belgium 7 2 Zebrafishlab, Veterinary Physiology and Biochemistry, Department of Veterinary 8 Sciences, University of Antwerp, Universiteitsplein 1, 2610 Wilrijk, Belgium 9 3 Systemic Physiological & Ecotoxicological Research, Department of Biology, 10 University of Antwerp, Universiteitsplein 1, 2610 Wilrijk, Belgium 11 4 Department of Biological Sciences and Center for Zebrafish Research, University of 12 Notre Dame, Notre Dame, -

Substitution of Isoleucine for Threonine at Position 190 of S-Opsin Causes S-Cone-Function Abnormalities ⇑ Rigmor C
Vision Research 73 (2012) 1–9 Contents lists available at SciVerse ScienceDirect Vision Research journal homepage: www.elsevier.com/locate/visres Substitution of isoleucine for threonine at position 190 of S-opsin causes S-cone-function abnormalities ⇑ Rigmor C. Baraas a, , Lene A. Hagen a, Elise W. Dees a, Maureen Neitz b a Department of Optometry & Visual Science, Buskerud University College, 3603 Kongsberg, Norway b Department of Ophthalmology, University of Washington Medical School, Seattle, WA 98195, USA article info abstract Article history: Five mutations in the S-cone–opsin gene (OPN1SW) that give rise to different single amino-acid substi- Received 15 October 2011 tutions (L56P, G79R, S214P, P264S, R283Q) are known to be associated with tritan color-vision deficiency. Received in revised form 30 August 2012 Here we report a sixth OPN1SW mutation (T190I) and the associated color vision phenotype. S-opsin Available online 26 September 2012 genotyping and clinical evaluation of color vision were performed on affected and unaffected family members and normal controls. Chromatic contrast was tested at different levels of retinal illuminance. Keywords: Affected family members were heterozygous for a nucleotide change that substituted the amino acid iso- Tritan color-vision deficiency leucine (I) in place of threonine (T) that is normally present at position 190 of the S-opsin. The mutation is Rod–cone interaction in extracellular loop II (EII). The association between making tritan errors and having the T190I mutant S S-opsin mutation opsin was strong (p > 0.0001: Fisher’s exact test). The performance of subjects with the T190I mutation was significantly different from that of normal trichromats along the tritan vector under all conditions tested (Mann–Whitney U: p < 0.05), but not along the protan or deutan vectors.