Aquatic Microbial Ecology 77:23
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evidence for Viral Infection in the Copepods Labidocera Aestiva And
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School January 2012 Evidence for Viral Infection in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida Darren Stephenson Dunlap University of South Florida, [email protected] Follow this and additional works at: http://scholarcommons.usf.edu/etd Part of the American Studies Commons, Other Oceanography and Atmospheric Sciences and Meteorology Commons, and the Virology Commons Scholar Commons Citation Dunlap, Darren Stephenson, "Evidence for Viral Infection in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida" (2012). Graduate Theses and Dissertations. http://scholarcommons.usf.edu/etd/4032 This Thesis is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Evidence of Viruses in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida By Darren S. Dunlap A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science College of Marine Science University of South Florida Major Professor: Mya Breitbart, Ph.D Kendra Daly, Ph.D Ian Hewson, Ph.D Date of Approval: March 19, 2012 Key Words: Copepods, Single-stranded DNA Viruses, Mesozooplankton, Transmission Electron Microscopy, Metagenomics Copyright © 2012, Darren Stephenson Dunlap DEDICATION None of this would have been possible without the generous love and support of my entire family over the years. My parents, Steve and Jill Dunlap, have always encouraged my pursuits with support and love, and their persistence of throwing me into lakes and rivers is largely responsible for my passion for Marine Science. -

Frequent Occurrence of Mungbean Yellow Mosaic India Virus in Tomato Leaf Curl Disease Afected Tomato in Oman M
www.nature.com/scientificreports OPEN Frequent occurrence of Mungbean yellow mosaic India virus in tomato leaf curl disease afected tomato in Oman M. S. Shahid 1*, M. Shafq 1, M. Ilyas2, A. Raza1, M. N. Al-Sadrani1, A. M. Al-Sadi 1 & R. W. Briddon 3 Next generation sequencing (NGS) of DNAs amplifed by rolling circle amplifcation from 6 tomato (Solanum lycopersicum) plants with leaf curl symptoms identifed a number of monopartite begomoviruses, including Tomato yellow leaf curl virus (TYLCV), and a betasatellite (Tomato leaf curl betasatellite [ToLCB]). Both TYLCV and ToLCB have previously been identifed infecting tomato in Oman. Surprisingly the NGS results also suggested the presence of the bipartite, legume-adapted begomovirus Mungbean yellow mosaic Indian virus (MYMIV). The presence of MYMIV was confrmed by cloning and Sanger sequencing from four of the six plants. A wider analysis by PCR showed MYMIV infection of tomato in Oman to be widespread. Inoculation of plants with full-length clones showed the host range of MYMIV not to extend to Nicotiana benthamiana or tomato. Inoculation to N. benthamiana showed TYLCV to be capable of maintaining MYMIV in both the presence and absence of the betasatellite. In tomato MYMIV was only maintained by TYLCV in the presence of the betasatellite and then only at low titre and efciency. This is the frst identifcation of TYLCV with ToLCB and the legume adapted bipartite begomovirus MYMIV co-infecting tomato. This fnding has far reaching implications. TYLCV has spread around the World from its origins in the Mediterranean/Middle East, in some instances, in live tomato planting material. -
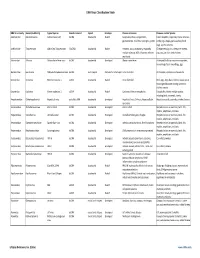
Virus Classification Tables V2.Vd.Xlsx
DNA Virus Classification Table DNA Virus Family Genera (Subfamily) Typical Species Genetic material Capsid Envelope Disease in Humans Diseases in other Species Adenoviridae Mastadenovirus Adenoviruses 1‐47 dsDNA Icosahedral Naked Respiratory illness; conjunctivitis, Canine hepatitis, respiratory illness in horses, gastroenteritis, tonsillitis, meningitis, cystitis cattle, pigs, sheep, goats, sea lions, birds dogs, squirrel enteritis Anelloviridae Torqueviruses Alpha‐Zeta Torqueviruses (‐)ssDNA Icosahedral Naked Hepatitis, lupus, pulmonary, myopathy, Chimpanzee, pig, cow, sheep, tree shrews, multiple sclerosis; 90% of humans infected pigs, cats, sea lions and chickens worldwide Asfarviridae Asfivirus African Swine fever virus dsDNA Icosahedral Enveloped African swine fever Arthropod (tick) transmission or ingestion; hemorrhagic fever in warthogs, pigs Baculoviridae Baculovirus Alpha‐Gamma Baculoviruses dsDNA Stick shaped Occluded or Enveloped none identified Arthropods, Lepidoptera, crustaceans Circoviridae Circovirus Porcine circovirus 1 ssDNA Icosahedral Naked none identified Birds, pigs, dogs; bats; rodents; causes post‐ weaning multisystem wasting syndrome, chicken anemia Circoviridae Cyclovirus Human cyclovirus 1 ssDNA Icosahedral Naked Cyclovirus Vietnam encephalitis Encephalitis; infects multiple species including birds, mammals, insects Hepadnaviridae Orthohepadnavirus Hepatitis B virus partially ssDNA Icosahedral Enveloped Hepatitis B virus; Cirrhosis, Hepatocellular Hepatitis in ducks, squirrels, primates, herons carcinoma Herpesviridae -

Viruses in Transplantation - Not Always Enemies
Viruses in transplantation - not always enemies Virome and transplantation ECCMID 2018 - Madrid Prof. Laurent Kaiser Head Division of Infectious Diseases Laboratory of Virology Geneva Center for Emerging Viral Diseases University Hospital of Geneva ESCMID eLibrary © by author Conflict of interest None ESCMID eLibrary © by author The human virome: definition? Repertoire of viruses found on the surface of/inside any body fluid/tissue • Eukaryotic DNA and RNA viruses • Prokaryotic DNA and RNA viruses (phages) 25 • The “main” viral community (up to 10 bacteriophages in humans) Haynes M. 2011, Metagenomic of the human body • Endogenous viral elements integrated into host chromosomes (8% of the human genome) • NGS is shaping the definition Rascovan N et al. Annu Rev Microbiol 2016;70:125-41 Popgeorgiev N et al. Intervirology 2013;56:395-412 Norman JM et al. Cell 2015;160:447-60 ESCMID eLibraryFoxman EF et al. Nat Rev Microbiol 2011;9:254-64 © by author Viruses routinely known to cause diseases (non exhaustive) Upper resp./oropharyngeal HSV 1 Influenza CNS Mumps virus Rhinovirus JC virus RSV Eye Herpes viruses Parainfluenza HSV Measles Coronavirus Adenovirus LCM virus Cytomegalovirus Flaviviruses Rabies HHV6 Poliovirus Heart Lower respiratory HTLV-1 Coxsackie B virus Rhinoviruses Parainfluenza virus HIV Coronaviruses Respiratory syncytial virus Parainfluenza virus Adenovirus Respiratory syncytial virus Coronaviruses Gastro-intestinal Influenza virus type A and B Human Bocavirus 1 Adenovirus Hepatitis virus type A, B, C, D, E Those that cause -

Identification of a Nanovirus-Alphasatellite Complex in Sophora Alopecuroides
Accepted Manuscript Title: Identification of a nanovirus-alphasatellite complex in Sophora alopecuroides Author: Jahangir Heydarnejad Mehdi Kamali Hossain Massumi Anders Kvarnheden Maketalena F. Male Simona Kraberger Daisy Stainton Darren P. Martin Arvind Varsani PII: S0168-1702(17)30009-6 DOI: http://dx.doi.org/doi:10.1016/j.virusres.2017.03.023 Reference: VIRUS 97113 To appear in: Virus Research Received date: 5-1-2017 Revised date: 15-3-2017 Accepted date: 18-3-2017 Please cite this article as: Heydarnejad, J., Kamali, M., Massumi, H., Kvarnheden, A., Male, M.F., Kraberger, S., Stainton, D., Martin, D.P., Varsani, A.,Identification of a nanovirus-alphasatellite complex in Sophora alopecuroides, Virus Research (2017), http://dx.doi.org/10.1016/j.virusres.2017.03.023 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. 1 Identification of a nanovirus-alphasatellite complex in Sophora alopecuroides 2 Jahangir Heydarnejad 1* , Mehdi Kamali 1, Hossain Massumi 1, Anders Kvarnheden 2, Maketalena 3 F. Male 3, Simona Kraberger 3,4 , Daisy Stainton 3,5 , Darren P. Martin 6 and Arvind Varsani 3,7,8* 4 5 1Department of Plant Protection, College -

Diversity and Evolution of Novel Invertebrate DNA Viruses Revealed by Meta-Transcriptomics
viruses Article Diversity and Evolution of Novel Invertebrate DNA Viruses Revealed by Meta-Transcriptomics Ashleigh F. Porter 1, Mang Shi 1, John-Sebastian Eden 1,2 , Yong-Zhen Zhang 3,4 and Edward C. Holmes 1,3,* 1 Marie Bashir Institute for Infectious Diseases and Biosecurity, Charles Perkins Centre, School of Life & Environmental Sciences and Sydney Medical School, The University of Sydney, Sydney, NSW 2006, Australia; [email protected] (A.F.P.); [email protected] (M.S.); [email protected] (J.-S.E.) 2 Centre for Virus Research, Westmead Institute for Medical Research, Westmead, NSW 2145, Australia 3 Shanghai Public Health Clinical Center and School of Public Health, Fudan University, Shanghai 201500, China; [email protected] 4 Department of Zoonosis, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Changping, Beijing 102206, China * Correspondence: [email protected]; Tel.: +61-2-9351-5591 Received: 17 October 2019; Accepted: 23 November 2019; Published: 25 November 2019 Abstract: DNA viruses comprise a wide array of genome structures and infect diverse host species. To date, most studies of DNA viruses have focused on those with the strongest disease associations. Accordingly, there has been a marked lack of sampling of DNA viruses from invertebrates. Bulk RNA sequencing has resulted in the discovery of a myriad of novel RNA viruses, and herein we used this methodology to identify actively transcribing DNA viruses in meta-transcriptomic libraries of diverse invertebrate species. Our analysis revealed high levels of phylogenetic diversity in DNA viruses, including 13 species from the Parvoviridae, Circoviridae, and Genomoviridae families of single-stranded DNA virus families, and six double-stranded DNA virus species from the Nudiviridae, Polyomaviridae, and Herpesviridae, for which few invertebrate viruses have been identified to date. -

An Unusual Alphasatellite Associated with Monopartite Begomoviruses Attenuates Symptoms and Reduces Betasatellite Accumulation
Journal of General Virology (2011), 92, 706–717 DOI 10.1099/vir.0.025288-0 An unusual alphasatellite associated with monopartite begomoviruses attenuates symptoms and reduces betasatellite accumulation Ali M. Idris,1,2 M. Shafiq Shahid,1,3 Rob W. Briddon,3 A. J. Khan,4 J.-K. Zhu2 and J. K. Brown1 Correspondence 1School of Plant Sciences, The University of Arizona, Tucson, AZ 85721, USA J. K. Brown 2Plant Stress Genomics Research Center, King Abdullah University of Science and Technology, [email protected] Thuwal 23955-6900, Kingdom of Saudi Arabia 3National Institute for Biotechnology and Genetic Engineering, PO Box 577, Jhang Road, Faisalabad, Pakistan 4Department of Crop Sciences, College of Agricultural and Marine Sciences, Sultan Qaboos University, PO Box 34, Al-Khod 123, Muscat, Sultanate of Oman The Oman strain of Tomato yellow leaf curl virus (TYLCV-OM) and its associated betasatellite, an isolate of Tomato leaf curl betasatellite (ToLCB), were previously reported from Oman. Here we report the isolation of a second, previously undescribed, begomovirus [Tomato leaf curl Oman virus (ToLCOMV)] and an alphasatellite from that same plant sample. This alphasatellite is closely related (90 % shared nucleotide identity) to an unusual DNA-2-type Ageratum yellow vein Singapore alphasatellite (AYVSGA), thus far identified only in Singapore. ToLCOMV was found to have a recombinant genome comprising sequences derived from two extant parents, TYLCV-OM, which is indigenous to Oman, and Papaya leaf curl virus from the Indian subcontinent. All possible combinations of ToLCOMV, TYLCV-OM, ToLCB and AYVSGA were used to agro-inoculate tomato and Nicotiana benthamiana. Infection with ToLCOMV yielded mild leaf-curl symptoms in both hosts; however, plants inoculated with TYLCV-OM developed more severe symptoms. -

The Intestinal Virome of Malabsorption Syndrome-Affected and Unaffected
Virus Research 261 (2019) 9–20 Contents lists available at ScienceDirect Virus Research journal homepage: www.elsevier.com/locate/virusres The intestinal virome of malabsorption syndrome-affected and unaffected broilers through shotgun metagenomics T ⁎ Diane A. Limaa, , Samuel P. Cibulskib, Caroline Tochettoa, Ana Paula M. Varelaa, Fabrine Finklera, Thais F. Teixeiraa, Márcia R. Loikoa, Cristine Cervac, Dennis M. Junqueirad, Fabiana Q. Mayerc, Paulo M. Roehea a Laboratório de Virologia, Departamento de Microbiologia, Imunologia e Parasitologia, Instituto de Ciências Básicas da Saúde (ICBS), Universidade Federal do Rio Grande do Sul (UFRGS), Porto Alegre, RS, Brazil b Laboratório de Virologia, Faculdade de Veterinária, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brazil c Laboratório de Biologia Molecular, Instituto de Pesquisas Veterinárias Desidério Finamor (IPVDF), Eldorado do Sul, RS, Brazil d Centro Universitário Ritter dos Reis - UniRitter, Health Science Department, Porto Alegre, RS, Brazil ARTICLE INFO ABSTRACT Keywords: Malabsorption syndrome (MAS) is an economically important disease of young, commercially reared broilers, Enteric disorders characterized by growth retardation, defective feather development and diarrheic faeces. Several viruses have Virome been tentatively associated to such syndrome. Here, in order to examine potential associations between enteric Broiler chickens viruses and MAS, the faecal viromes of 70 stool samples collected from diseased (n = 35) and healthy (n = 35) High-throughput sequencing chickens from seven flocks were characterized and compared. Following high-throughput sequencing, a total of 8,347,319 paired end reads, with an average of 231 nt, were generated. Through analysis of de novo assembled contigs, 144 contigs > 1000 nt were identified with hits to eukaryotic viral sequences, as determined by GenBank database. -

Single Stranded DNA Viruses Associated with Capybara Faeces Sampled in Brazil
viruses Article Single Stranded DNA Viruses Associated with Capybara Faeces Sampled in Brazil Rafaela S. Fontenele 1,2, Cristiano Lacorte 2, Natalia S. Lamas 2, Kara Schmidlin 1, Arvind Varsani 1,3,* and Simone G. Ribeiro 2,* 1 The Biodesign Center for Fundamental and Applied Microbiomics, Center for Evolution and Medicine School of Life Sciences, Arizona State University, Tempe, AZ 85287, USA 2 Embrapa Recursos Genéticos e Biotecnologia, Brasília, DF 70770-017, Brazil 3 Structural Biology Research Unit, Department of Clinical Laboratory Sciences, University of Cape Town, Observatory, Cape Town 7925, South Africa * Correspondence: [email protected] (A.V.); [email protected] (S.G.R.); Tel.: +1-480-727-2093 (A.V.); +55-61-3448-4666/3448-4703 (S.G.R.) Received: 14 June 2019; Accepted: 31 July 2019; Published: 2 August 2019 Abstract: Capybaras (Hydrochoerus hydrochaeris), the world’s largest rodents, are distributed throughout South America. These wild herbivores are commonly found near water bodies and are well adapted to rural and urban areas. There is limited information on the viruses circulating through capybaras. This study aimed to expand the knowledge on the viral diversity associated with capybaras by sampling their faeces. Using a viral metagenomics approach, we identified diverse single-stranded DNA viruses in the capybara faeces sampled in the Distrito Federal, Brazil. A total of 148 complete genomes of viruses in the Microviridae family were identified. In addition, 14 genomoviruses (family Genomoviridae), a novel cyclovirus (family Circoviridae), and a smacovirus (family Smacoviridae) were identified. Also, 37 diverse viruses that cannot be assigned to known families and more broadly referred to as unclassified circular replication associated protein encoding single-stranded (CRESS) DNA viruses were identified. -

The Discovery, Distribution and Diversity of DNA Viruses
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.16.342956; this version posted March 17, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Title: The discovery, distribution and diversity of DNA viruses associated with Drosophila melanogaster in Europe Running title: DNA viruses of European Drosophila Key Words: DNA virus, Endogenous viral element, Drosophila, Nudivirus, Galbut virus, Filamentous virus, Adintovirus, Densovirus, Bidnavirus Authors: Megan A. Wallace 1,2 [email protected] 0000-0001-5367-420X Kelsey A. Coffman 3 [email protected] 0000-0002-7609-6286 Clément Gilbert 1,4 [email protected] 0000-0002-2131-7467 Sanjana Ravindran 2 [email protected] 0000-0003-0996-0262 Gregory F. Albery 5 [email protected] 0000-0001-6260-2662 Jessica Abbott 1,6 [email protected] 0000-0002-8743-2089 Eliza Argyridou 1,7 [email protected] 0000-0002-6890-4642 Paola Bellosta 1,8,9 [email protected] 0000-0003-1913-5661 Andrea J. Betancourt 1,10 [email protected] 0000-0001-9351-1413 Hervé Colinet 1,11 [email protected] 0000-0002-8806-3107 Katarina Eric 1,12 [email protected] 0000-0002-3456-2576 Amanda Glaser-Schmitt 1,7 [email protected] 0000-0002-1322-1000 Sonja Grath 1,7 [email protected] 0000-0003-3621-736X Mihailo Jelic 1,13 [email protected] 0000-0002-1637-0933 Maaria Kankare 1,14 [email protected] 0000-0003-1541-9050 Iryna Kozeretska 1,15 [email protected] 0000-0002-6485-1408 Volker Loeschcke 1,16 [email protected] 0000-0003-1450-0754 Catherine Montchamp-Moreau 1,4 [email protected] 0000-0002-5044-9709 Lino Ometto 1,17 [email protected] 0000-0002-2679-625X Banu Sebnem Onder 1,18 [email protected] 0000-0002-3003-248X Dorcas J. -

Intestinal Virome Changes Precede Autoimmunity in Type I Diabetes-Susceptible Children,” by Guoyan Zhao, Tommi Vatanen, Lindsay Droit, Arnold Park, Aleksandar D
Correction MEDICAL SCIENCES Correction for “Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children,” by Guoyan Zhao, Tommi Vatanen, Lindsay Droit, Arnold Park, Aleksandar D. Kostic, Tiffany W. Poon, Hera Vlamakis, Heli Siljander, Taina Härkönen, Anu-Maaria Hämäläinen, Aleksandr Peet, Vallo Tillmann, Jorma Ilonen, David Wang, Mikael Knip, Ramnik J. Xavier, and Herbert W. Virgin, which was first published July 10, 2017; 10.1073/pnas.1706359114 (Proc Natl Acad Sci USA 114: E6166–E6175). The authors wish to note the following: “After publication, we discovered that certain patient-related information in the spreadsheets placed online had information that could conceiv- ably be used to identify, or at least narrow down, the identity of children whose fecal samples were studied. The article has been updated online to remove these potential privacy concerns. These changes do not alter the conclusions of the paper.” Published under the PNAS license. Published online November 19, 2018. www.pnas.org/cgi/doi/10.1073/pnas.1817913115 E11426 | PNAS | November 27, 2018 | vol. 115 | no. 48 www.pnas.org Downloaded by guest on September 26, 2021 Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children Guoyan Zhaoa,1, Tommi Vatanenb,c, Lindsay Droita, Arnold Parka, Aleksandar D. Kosticb,2, Tiffany W. Poonb, Hera Vlamakisb, Heli Siljanderd,e, Taina Härkönend,e, Anu-Maaria Hämäläinenf, Aleksandr Peetg,h, Vallo Tillmanng,h, Jorma Iloneni, David Wanga,j, Mikael Knipd,e,k,l, Ramnik J. Xavierb,m, and -

Suivi De La Thèse
UNIVERSITE OUAGA I UNIVERSITÉ DE LA PR JOSEPH KI-ZERBO RÉUNION ---------- ---------- Unité de Formation et de Recherche Faculté des Sciences et Sciences de la Vie et de la Terre Technologies UMR Peuplements Végétaux et Bio-agresseurs en Milieu Tropical CIRAD – Université de La Réunion INERA – LMI Patho-Bios THÈSE EN COTUTELLE Pour obtenir le diplôme de Doctorat en Sciences Epidémiologie moléculaire des géminivirus responsables de maladies émergentes sur les cultures maraîchères au Burkina Faso par Alassane Ouattara Soutenance le 14 Décembre 2017, devant le jury composé de Stéphane POUSSIER Professeur, Université de La Réunion Président Justin PITA Professeur, Université Houphouët-Boigny, Côte d'Ivoire Rapporteur Philippe ROUMAGNAC Chercheur HDR, CIRAD, UMR BGPI, France Rapporteur Fidèle TIENDREBEOGO Chercheur, INERA, Burkina Faso Examinateur Nathalie BECKER Maître de conférences HDR MNHN, UMR ISYEB, France Examinatrice Jean-Michel LETT Chercheur HDR, CIRAD, UMR PVBMT, La Réunion Co-Directeur de thèse Nicolas BARRO Professeur, Université Ouagadougou, Burkina Faso Co-Directeur de thèse DEDICACES A mon épouse Dadjata et à mon fils Jaad Kaamil: merci pour l’amour, les encouragements et la compréhension tout au long de ces trois années. A mon père Kassoum, à ma mère Salimata, à ma belle-mère Bila, à mon oncle Soumaïla et son épouse Abibata : merci pour votre amour, j’ai toujours reçu soutien, encouragements et bénédictions de votre part. Puisse Dieu vous garder en bonne santé ! REMERCIEMENTS Mes remerciements vont à l’endroit du personnel des Universités Ouaga I Pr Joseph KI- ZERBO et de La Réunion pour avoir accepté mon inscription. Je remercie les différents financeurs de mes travaux : l’AIRD (Projet PEERS-EMEB), l’Union Européenne (ERDF), le Conseil Régional de La Réunion et le Cirad (Bourse Cirad-Sud).