An Unconventional Flavivirus and Other RNA Viruses In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Petition to List the Black Teatfish, Holothuria Nobilis, Under the U.S. Endangered Species Act
Before the Secretary of Commerce Petition to List the Black Teatfish, Holothuria nobilis, under the U.S. Endangered Species Act Photo Credit: © Philippe Bourjon (with permission) Center for Biological Diversity 14 May 2020 Notice of Petition Wilbur Ross, Secretary of Commerce U.S. Department of Commerce 1401 Constitution Ave. NW Washington, D.C. 20230 Email: [email protected], [email protected] Dr. Neil Jacobs, Acting Under Secretary of Commerce for Oceans and Atmosphere U.S. Department of Commerce 1401 Constitution Ave. NW Washington, D.C. 20230 Email: [email protected] Petitioner: Kristin Carden, Oceans Program Scientist Sarah Uhlemann, Senior Att’y & Int’l Program Director Center for Biological Diversity Center for Biological Diversity 1212 Broadway #800 2400 NW 80th Street, #146 Oakland, CA 94612 Seattle,WA98117 Phone: (510) 844‐7100 x327 Phone: (206) 324‐2344 Email: [email protected] Email: [email protected] The Center for Biological Diversity (Center, Petitioner) submits to the Secretary of Commerce and the National Oceanographic and Atmospheric Administration (NOAA) through the National Marine Fisheries Service (NMFS) a petition to list the black teatfish, Holothuria nobilis, as threatened or endangered under the U.S. Endangered Species Act (ESA), 16 U.S.C. § 1531 et seq. Alternatively, the Service should list the black teatfish as threatened or endangered throughout a significant portion of its range. This species is found exclusively in foreign waters, thus 30‐days’ notice to affected U.S. states and/or territories was not required. The Center is a non‐profit, public interest environmental organization dedicated to the protection of native species and their habitats. -

On the Entry and Entrapment of Picornaviruses
On the entry and entrapment of picornaviruses Jacqueline Staring PS_JS_def.indd 1 08-10-18 08:46 ISBN 978-94-92679-60-4 NUR 100 Printing and lay-out by: Proefschriftenprinten.nl – The Netherlands With financial support from the Netherlands Cancer Institute © J. Staring, 2018 All rights are reserved. No part of this book may be reproduced, distributed, or transmitted in any form or by any means, without prior written permission of the author. PS_JS_def.indd 2 08-10-18 08:46 On the entry and entrapment of picornaviruses Over het binnendringen en insluiten van picornavirussen (met een samenvatting in het Nederlands) Proefschrift ter verkrijging van de graad van doctor aan de Universiteit Utrecht op gezag van de rector magnificus, prof. dr. H.R.B.M. Kummeling, ingevolge het besluit van het college voor promoties in het openbaar te verdedigen op dinsdag 6 november 2018 des middags te 16.15 uur door Jacqueline Staring geboren op 8 december 1984 te Leiden PS_JS_def.indd 3 08-10-18 08:46 Promotor: Prof. dr T.R. Brummelkamp PS_JS_def.indd 4 08-10-18 08:46 TABLE OF CONTENTS Chapter 1 Introduction I: Picornaviruses 7 Chapter 2 Introduction II: Genetic approaches to study 27 host-pathogen interactions Chapter 3 PLA2G16, a switch between entry and clearance of 43 picornaviridae Chapter 4 Enterovirus D68 receptor requirements unveiled by 77 haploid genetics Chapter 5 KREMEN1 is a host entry receptor for a major group 95 of enteroviruses Chapter 6 General Discussion I: Viral endosomal escape & 125 detection at a glance Chapter 7 General Discussion II: Concluding remarks 151 Addendum 161 Summary 163 Nederlandse samenvatting 165 Curriculum vitae 167 List of publications 168 Acknowledgements 169 PS_JS_def.indd 5 08-10-18 08:46 Roll the dice if you’re going to try, go all the way. -

Evidence for Viral Infection in the Copepods Labidocera Aestiva And
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School January 2012 Evidence for Viral Infection in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida Darren Stephenson Dunlap University of South Florida, [email protected] Follow this and additional works at: http://scholarcommons.usf.edu/etd Part of the American Studies Commons, Other Oceanography and Atmospheric Sciences and Meteorology Commons, and the Virology Commons Scholar Commons Citation Dunlap, Darren Stephenson, "Evidence for Viral Infection in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida" (2012). Graduate Theses and Dissertations. http://scholarcommons.usf.edu/etd/4032 This Thesis is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Evidence of Viruses in the Copepods Labidocera aestiva and Acartia tonsa in Tampa Bay, Florida By Darren S. Dunlap A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science College of Marine Science University of South Florida Major Professor: Mya Breitbart, Ph.D Kendra Daly, Ph.D Ian Hewson, Ph.D Date of Approval: March 19, 2012 Key Words: Copepods, Single-stranded DNA Viruses, Mesozooplankton, Transmission Electron Microscopy, Metagenomics Copyright © 2012, Darren Stephenson Dunlap DEDICATION None of this would have been possible without the generous love and support of my entire family over the years. My parents, Steve and Jill Dunlap, have always encouraged my pursuits with support and love, and their persistence of throwing me into lakes and rivers is largely responsible for my passion for Marine Science. -

Coordinated Action of RTBV and RTSV Proteins Suppress Host RNA Silencing Machinery
bioRxiv preprint doi: https://doi.org/10.1101/2021.01.19.427099; this version posted January 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Coordinated action of RTBV and RTSV proteins suppress host RNA silencing machinery Abhishek Anand1, Malathi Pinninti2, Anita Tripathi1, Satendra Kumar Mangrauthia2 and Neeti Sanan-Mishra1* 1 Plant RNAi Biology Group, International Center for Genetic Engineering and Biotechnology, New Delhi-110067 2 Biotechnology Section, ICAR-Indian Institute of Rice Research, Rajendranangar, Hyderabad- 500030 *Corresponding Author: Neeti Sanan-Mishra E-mail address: [email protected] Author e-mail: Abhishek Anand: [email protected] Malathi Pinninti: [email protected] Anita Tripathi: [email protected] Satendra K. Mangrauthia: [email protected] Abstract RNA silencing is as an adaptive immune response in plants that limits accumulation or spread of invading viruses. Successful virus infection entails countering the RNA silencing for efficient replication and systemic spread in the host. The viruses encode proteins having the ability to suppress or block the host silencing mechanism, resulting in severe pathogenic symptoms and diseases. Tungro virus disease caused by a complex of two viruses provides an excellent system to understand these host and virus interactions during infection. It is known that Rice tungro bacilliform virus (RTBV) is the major determinant of the disease while Rice tungro spherical virus (RTSV) accentuates the symptoms. This study brings to focus the important role of RTBV ORF-IV in Tungro disease manifestation, by acting as both the victim and silencer of the RNA silencing pathway. -

Marine Invertebrate Field Guide
Marine Invertebrate Field Guide Contents ANEMONES ....................................................................................................................................................................................... 2 AGGREGATING ANEMONE (ANTHOPLEURA ELEGANTISSIMA) ............................................................................................................................... 2 BROODING ANEMONE (EPIACTIS PROLIFERA) ................................................................................................................................................... 2 CHRISTMAS ANEMONE (URTICINA CRASSICORNIS) ............................................................................................................................................ 3 PLUMOSE ANEMONE (METRIDIUM SENILE) ..................................................................................................................................................... 3 BARNACLES ....................................................................................................................................................................................... 4 ACORN BARNACLE (BALANUS GLANDULA) ....................................................................................................................................................... 4 HAYSTACK BARNACLE (SEMIBALANUS CARIOSUS) .............................................................................................................................................. 4 CHITONS ........................................................................................................................................................................................... -

Multiple Origins of Prokaryotic and Eukaryotic Single-Stranded DNA Viruses from Bacterial and Archaeal Plasmids
ARTICLE https://doi.org/10.1038/s41467-019-11433-0 OPEN Multiple origins of prokaryotic and eukaryotic single-stranded DNA viruses from bacterial and archaeal plasmids Darius Kazlauskas 1, Arvind Varsani 2,3, Eugene V. Koonin 4 & Mart Krupovic 5 Single-stranded (ss) DNA viruses are a major component of the earth virome. In particular, the circular, Rep-encoding ssDNA (CRESS-DNA) viruses show high diversity and abundance 1234567890():,; in various habitats. By combining sequence similarity network and phylogenetic analyses of the replication proteins (Rep) belonging to the HUH endonuclease superfamily, we show that the replication machinery of the CRESS-DNA viruses evolved, on three independent occa- sions, from the Reps of bacterial rolling circle-replicating plasmids. The CRESS-DNA viruses emerged via recombination between such plasmids and cDNA copies of capsid genes of eukaryotic positive-sense RNA viruses. Similarly, the rep genes of prokaryotic DNA viruses appear to have evolved from HUH endonuclease genes of various bacterial and archaeal plasmids. Our findings also suggest that eukaryotic polyomaviruses and papillomaviruses with dsDNA genomes have evolved via parvoviruses from CRESS-DNA viruses. Collectively, our results shed light on the complex evolutionary history of a major class of viruses revealing its polyphyletic origins. 1 Institute of Biotechnology, Life Sciences Center, Vilnius University, Saulėtekio av. 7, Vilnius 10257, Lithuania. 2 The Biodesign Center for Fundamental and Applied Microbiomics, School of Life Sciences, Center for Evolution and Medicine, Arizona State University, Tempe, AZ 85287, USA. 3 Structural Biology Research Unit, Department of Integrative Biomedical Sciences, University of Cape Town, Rondebosch, 7700 Cape Town, South Africa. -

Origin, Adaptation and Evolutionary Pathways of Fungal Viruses
Virus Genes 16:1, 119±131, 1998 # 1998 Kluwer Academic Publishers, Boston. Manufactured in The Netherlands. Origin, Adaptation and Evolutionary Pathways of Fungal Viruses SAID A. GHABRIAL Department of Plant Pathology, University of Kentucky, Lexington, KY, USA Abstract. Fungal viruses or mycoviruses are widespread in fungi and are believed to be of ancient origin. They have evolved in concert with their hosts and are usually associated with symptomless infections. Mycoviruses are transmitted intracellularly during cell division, sporogenesis and cell fusion, and they lack an extracellular phase to their life cycles. Their natural host ranges are limited to individuals within the same or closely related vegetative compatibility groups. Typically, fungal viruses are isometric particles 25±50 nm in diameter, and possess dsRNA genomes. The best characterized of these belong to the family Totiviridae whose members have simple undivided dsRNA genomes comprised of a coat protein (CP) gene and an RNA dependent RNA polymerase (RDRP) gene. A recently characterized totivirus infecting a ®lamentous fungus was found to be more closely related to protozoan totiviruses than to yeast totiviruses suggesting these viruses existed prior to the divergence of fungi and protozoa. Although the dsRNA viruses at large are polyphyletic, based on RDRP sequence comparisons, the totiviruses are monophyletic. The theory of a cellular self-replicating mRNA as the origin of totiviruses is attractive because of their apparent ancient origin, the close relationships among their RDRPs, genome simplicity and the ability to use host proteins ef®ciently. Mycoviruses with bipartite genomes ( partitiviruses), like the totiviruses, have simple genomes, but the CP and RDRP genes are on separate dsRNA segments. -

2020 Taxonomic Update for Phylum Negarnaviricota (Riboviria: Orthornavirae), Including the Large Orders Bunyavirales and Mononegavirales
Archives of Virology https://doi.org/10.1007/s00705-020-04731-2 VIROLOGY DIVISION NEWS 2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales Jens H. Kuhn1 · Scott Adkins2 · Daniela Alioto3 · Sergey V. Alkhovsky4 · Gaya K. Amarasinghe5 · Simon J. Anthony6,7 · Tatjana Avšič‑Županc8 · María A. Ayllón9,10 · Justin Bahl11 · Anne Balkema‑Buschmann12 · Matthew J. Ballinger13 · Tomáš Bartonička14 · Christopher Basler15 · Sina Bavari16 · Martin Beer17 · Dennis A. Bente18 · Éric Bergeron19 · Brian H. Bird20 · Carol Blair21 · Kim R. Blasdell22 · Steven B. Bradfute23 · Rachel Breyta24 · Thomas Briese25 · Paul A. Brown26 · Ursula J. Buchholz27 · Michael J. Buchmeier28 · Alexander Bukreyev18,29 · Felicity Burt30 · Nihal Buzkan31 · Charles H. Calisher32 · Mengji Cao33,34 · Inmaculada Casas35 · John Chamberlain36 · Kartik Chandran37 · Rémi N. Charrel38 · Biao Chen39 · Michela Chiumenti40 · Il‑Ryong Choi41 · J. Christopher S. Clegg42 · Ian Crozier43 · John V. da Graça44 · Elena Dal Bó45 · Alberto M. R. Dávila46 · Juan Carlos de la Torre47 · Xavier de Lamballerie38 · Rik L. de Swart48 · Patrick L. Di Bello49 · Nicholas Di Paola50 · Francesco Di Serio40 · Ralf G. Dietzgen51 · Michele Digiaro52 · Valerian V. Dolja53 · Olga Dolnik54 · Michael A. Drebot55 · Jan Felix Drexler56 · Ralf Dürrwald57 · Lucie Dufkova58 · William G. Dundon59 · W. Paul Duprex60 · John M. Dye50 · Andrew J. Easton61 · Hideki Ebihara62 · Toufc Elbeaino63 · Koray Ergünay64 · Jorlan Fernandes195 · Anthony R. Fooks65 · Pierre B. H. Formenty66 · Leonie F. Forth17 · Ron A. M. Fouchier48 · Juliana Freitas‑Astúa67 · Selma Gago‑Zachert68,69 · George Fú Gāo70 · María Laura García71 · Adolfo García‑Sastre72 · Aura R. Garrison50 · Aiah Gbakima73 · Tracey Goldstein74 · Jean‑Paul J. Gonzalez75,76 · Anthony Grifths77 · Martin H. Groschup12 · Stephan Günther78 · Alexandro Guterres195 · Roy A. -

SPC Beche-De-Mer Information Bulletin #39 – March 2019
ISSN 1025-4943 Issue 39 – March 2019 BECHE-DE-MER information bulletin v Inside this issue Editorial Towards producing a standard grade identification guide for bêche-de-mer in This issue of the Beche-de-mer Information Bulletin is well supplied with Solomon Islands 15 articles that address various aspects of the biology, fisheries and S. Lee et al. p. 3 aquaculture of sea cucumbers from three major oceans. An assessment of commercial sea cu- cumber populations in French Polynesia Lee and colleagues propose a procedure for writing guidelines for just after the 2012 moratorium the standard identification of beche-de-mer in Solomon Islands. S. Andréfouët et al. p. 8 Andréfouët and colleagues assess commercial sea cucumber Size at sexual maturity of the flower populations in French Polynesia and discuss several recommendations teatfish Holothuria (Microthele) sp. in the specific to the different archipelagos and islands, in the view of new Seychelles management decisions. Cahuzac and others studied the reproductive S. Cahuzac et al. p. 19 biology of Holothuria species on the Mahé and Amirantes plateaux Contribution to the knowledge of holo- in the Seychelles during the 2018 northwest monsoon season. thurian biodiversity at Reunion Island: Two previously unrecorded dendrochi- Bourjon and Quod provide a new contribution to the knowledge of rotid sea cucumbers species (Echinoder- holothurian biodiversity on La Réunion, with observations on two mata: Holothuroidea). species that are previously undescribed. Eeckhaut and colleagues P. Bourjon and J.-P. Quod p. 27 show that skin ulcerations of sea cucumbers in Madagascar are one Skin ulcerations in Holothuria scabra can symptom of different diseases induced by various abiotic or biotic be induced by various types of food agents. -

Research on Indian Echinoderms - a Review
J. mar. biol Ass. India, 1983, 25 (1 & 2):91 -108 RESEARCH ON INDIAN ECHINODERMS - A REVIEW D. B. JAMES* Central Marine fisheries Research Institute, Cochin -682031 In the present paper research work so far done on Indian Echinoderms is reviewed. Various aspects such as history, taxonomy, anatomy, reproductive physiology, development and larval forms, ecology, animal associations, parasites, utility, distribution and zoogeography, toxicology and bibliography are reviewed in detail. Corrections to misidentifications in earlier papers have been made wherever possible and presented in the Appendix at the end of the paper. and help at every stage. He thanks Dr. INTRODUCTION P.S.B.R. James, Director, C.M.F.R. Institute for kindly suggesting to write this review and ECHINODERMS being common and conspicuous also for his kind interest and encouragement. organisms of the sea shore have attracted the He also thanks Miss A.M. Clark, British Museum attention of the naturalists since very early (Natural History), London for commenting times. Their btauty, more so their symmetry on the correct identity of some of the echino have attracted the attention of many a natura derms presented here. list. Although the first paper on Indian Echino- <jlerms was published as early as 1743 by Plan- HISTORY cus and Gaultire from Goa there was not much progress in the field except in the late nineteenth Most of the research work on Indian echino and early twentieth centuries when echinoderms derms relate only to taxonomy with little or no collected mostly by R.I.M.S. Investigator were information on the ecology and other aspects reported by several authors. -

The Biology of Seashores - Image Bank Guide All Images and Text ©2006 Biomedia ASSOCIATES
The Biology of Seashores - Image Bank Guide All Images And Text ©2006 BioMEDIA ASSOCIATES Shore Types Low tide, sandy beach, clam diggers. Knowing the Low tide, rocky shore, sandstone shelves ,The time and extent of low tides is important for people amount of beach exposed at low tide depends both on who collect intertidal organisms for food. the level the tide will reach, and on the gradient of the beach. Low tide, Salt Point, CA, mixed sandstone and hard Low tide, granite boulders, The geology of intertidal rock boulders. A rocky beach at low tide. Rocks in the areas varies widely. Here, vertical faces of exposure background are about 15 ft. (4 meters) high. are mixed with gentle slopes, providing much variation in rocky intertidal habitat. Split frame, showing low tide and high tide from same view, Salt Point, California. Identical views Low tide, muddy bay, Bodega Bay, California. of a rocky intertidal area at a moderate low tide (left) Bays protected from winds, currents, and waves tend and moderate high tide (right). Tidal variation between to be shallow and muddy as sediments from rivers these two times was about 9 feet (2.7 m). accumulate in the basin. The receding tide leaves mudflats. High tide, Salt Point, mixed sandstone and hard rock boulders. Same beach as previous two slides, Low tide, muddy bay. In some bays, low tides expose note the absence of exposed algae on the rocks. vast areas of mudflats. The sea may recede several kilometers from the shoreline of high tide Tides Low tide, sandy beach. -
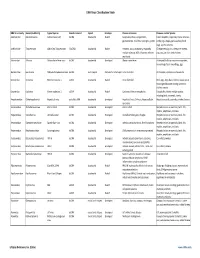
Virus Classification Tables V2.Vd.Xlsx
DNA Virus Classification Table DNA Virus Family Genera (Subfamily) Typical Species Genetic material Capsid Envelope Disease in Humans Diseases in other Species Adenoviridae Mastadenovirus Adenoviruses 1‐47 dsDNA Icosahedral Naked Respiratory illness; conjunctivitis, Canine hepatitis, respiratory illness in horses, gastroenteritis, tonsillitis, meningitis, cystitis cattle, pigs, sheep, goats, sea lions, birds dogs, squirrel enteritis Anelloviridae Torqueviruses Alpha‐Zeta Torqueviruses (‐)ssDNA Icosahedral Naked Hepatitis, lupus, pulmonary, myopathy, Chimpanzee, pig, cow, sheep, tree shrews, multiple sclerosis; 90% of humans infected pigs, cats, sea lions and chickens worldwide Asfarviridae Asfivirus African Swine fever virus dsDNA Icosahedral Enveloped African swine fever Arthropod (tick) transmission or ingestion; hemorrhagic fever in warthogs, pigs Baculoviridae Baculovirus Alpha‐Gamma Baculoviruses dsDNA Stick shaped Occluded or Enveloped none identified Arthropods, Lepidoptera, crustaceans Circoviridae Circovirus Porcine circovirus 1 ssDNA Icosahedral Naked none identified Birds, pigs, dogs; bats; rodents; causes post‐ weaning multisystem wasting syndrome, chicken anemia Circoviridae Cyclovirus Human cyclovirus 1 ssDNA Icosahedral Naked Cyclovirus Vietnam encephalitis Encephalitis; infects multiple species including birds, mammals, insects Hepadnaviridae Orthohepadnavirus Hepatitis B virus partially ssDNA Icosahedral Enveloped Hepatitis B virus; Cirrhosis, Hepatocellular Hepatitis in ducks, squirrels, primates, herons carcinoma Herpesviridae