Evidence for Viral Infection in the Copepods Labidocera Aestiva And
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Guide for Common Viral Diseases of Animals in Louisiana
Sampling and Testing Guide for Common Viral Diseases of Animals in Louisiana Please click on the species of interest: Cattle Deer and Small Ruminants The Louisiana Animal Swine Disease Diagnostic Horses Laboratory Dogs A service unit of the LSU School of Veterinary Medicine Adapted from Murphy, F.A., et al, Veterinary Virology, 3rd ed. Cats Academic Press, 1999. Compiled by Rob Poston Multi-species: Rabiesvirus DCN LADDL Guide for Common Viral Diseases v. B2 1 Cattle Please click on the principle system involvement Generalized viral diseases Respiratory viral diseases Enteric viral diseases Reproductive/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 2 Deer and Small Ruminants Please click on the principle system involvement Generalized viral disease Respiratory viral disease Enteric viral diseases Reproductive/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 3 Swine Please click on the principle system involvement Generalized viral diseases Respiratory viral diseases Enteric viral diseases Reproductive/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 4 Horses Please click on the principle system involvement Generalized viral diseases Neurological viral diseases Respiratory viral diseases Enteric viral diseases Abortifacient/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 5 Dogs Please click on the principle system involvement Generalized viral diseases Respiratory viral diseases Enteric viral diseases Reproductive/neonatal viral diseases Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. -

Virus–Host Interactions and Their Roles in Coral Reef Health and Disease
!"#$"%& Virus–host interactions and their roles in coral reef health and disease Rebecca Vega Thurber1, Jérôme P. Payet1,2, Andrew R. Thurber1,2 and Adrienne M. S. Correa3 !"#$%&'$()(*+%&,(%--.#(+''/%!01(1/$%0-1$23++%(#4&,,+5(5&$-%#6('+1#$0$/$-("0+708-%#0$9(&17( 3%+7/'$080$9(4+$#3+$#6(&17(&%-($4%-&$-1-7("9(&1$4%+3+:-10'(70#$/%"&1'-;(<40#(=-80-5(3%+807-#( &1(01$%+7/'$0+1($+('+%&,(%--.(80%+,+:9(&17(->34�?-#($4-(,01@#("-$5--1(80%/#-#6('+%&,(>+%$&,0$9( &17(%--.(-'+#9#$->(7-',01-;(A-(7-#'%0"-($4-(70#$01'$08-("-1$40'2&##+'0&$-7(&17(5&$-%2'+,/>12( &##+'0&$-7(80%+>-#($4&$(&%-(/10B/-($+('+%&,(%--.#6(540'4(4&8-(%-'-08-7(,-##(&$$-1$0+1($4&1( 80%/#-#(01(+3-12+'-&1(#9#$->#;(A-(493+$4-#0?-($4&$(80%/#-#(+.("&'$-%0&(&17(-/@&%9+$-#( 791&>0'&,,9(01$-%&'$(50$4($4-0%(4+#$#(01($4-(5&$-%('+,/>1(&17(50$4(#',-%&'$010&1(C#$+19D('+%&,#($+( 01.,/-1'-(>0'%+"0&,('+>>/10$9(791&>0'#6('+%&,(",-&'401:(&17(70#-&#-6(&17(%--.("0+:-+'4->0'&,( cycling. Last, we outline how marine viruses are an integral part of the reef system and suggest $4&$($4-(01.,/-1'-(+.(80%/#-#(+1(%--.(./1'$0+1(0#(&1(-##-1$0&,('+>3+1-1$(+.($4-#-(:,+"&,,9( 0>3+%$&1$(-180%+1>-1$#; To p - d ow n e f f e c t s Viruses infect all cellular life, including bacteria and evidence that macroorganisms play important parts in The ecological concept that eukaryotes, and contain ~200 megatonnes of carbon the dynamics of viroplankton; for example, sponges can organismal growth and globally1 — thus, they are integral parts of marine eco- filter and consume viruses6,7. -
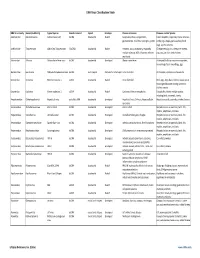
Virus Classification Tables V2.Vd.Xlsx
DNA Virus Classification Table DNA Virus Family Genera (Subfamily) Typical Species Genetic material Capsid Envelope Disease in Humans Diseases in other Species Adenoviridae Mastadenovirus Adenoviruses 1‐47 dsDNA Icosahedral Naked Respiratory illness; conjunctivitis, Canine hepatitis, respiratory illness in horses, gastroenteritis, tonsillitis, meningitis, cystitis cattle, pigs, sheep, goats, sea lions, birds dogs, squirrel enteritis Anelloviridae Torqueviruses Alpha‐Zeta Torqueviruses (‐)ssDNA Icosahedral Naked Hepatitis, lupus, pulmonary, myopathy, Chimpanzee, pig, cow, sheep, tree shrews, multiple sclerosis; 90% of humans infected pigs, cats, sea lions and chickens worldwide Asfarviridae Asfivirus African Swine fever virus dsDNA Icosahedral Enveloped African swine fever Arthropod (tick) transmission or ingestion; hemorrhagic fever in warthogs, pigs Baculoviridae Baculovirus Alpha‐Gamma Baculoviruses dsDNA Stick shaped Occluded or Enveloped none identified Arthropods, Lepidoptera, crustaceans Circoviridae Circovirus Porcine circovirus 1 ssDNA Icosahedral Naked none identified Birds, pigs, dogs; bats; rodents; causes post‐ weaning multisystem wasting syndrome, chicken anemia Circoviridae Cyclovirus Human cyclovirus 1 ssDNA Icosahedral Naked Cyclovirus Vietnam encephalitis Encephalitis; infects multiple species including birds, mammals, insects Hepadnaviridae Orthohepadnavirus Hepatitis B virus partially ssDNA Icosahedral Enveloped Hepatitis B virus; Cirrhosis, Hepatocellular Hepatitis in ducks, squirrels, primates, herons carcinoma Herpesviridae -

Viruses in Transplantation - Not Always Enemies
Viruses in transplantation - not always enemies Virome and transplantation ECCMID 2018 - Madrid Prof. Laurent Kaiser Head Division of Infectious Diseases Laboratory of Virology Geneva Center for Emerging Viral Diseases University Hospital of Geneva ESCMID eLibrary © by author Conflict of interest None ESCMID eLibrary © by author The human virome: definition? Repertoire of viruses found on the surface of/inside any body fluid/tissue • Eukaryotic DNA and RNA viruses • Prokaryotic DNA and RNA viruses (phages) 25 • The “main” viral community (up to 10 bacteriophages in humans) Haynes M. 2011, Metagenomic of the human body • Endogenous viral elements integrated into host chromosomes (8% of the human genome) • NGS is shaping the definition Rascovan N et al. Annu Rev Microbiol 2016;70:125-41 Popgeorgiev N et al. Intervirology 2013;56:395-412 Norman JM et al. Cell 2015;160:447-60 ESCMID eLibraryFoxman EF et al. Nat Rev Microbiol 2011;9:254-64 © by author Viruses routinely known to cause diseases (non exhaustive) Upper resp./oropharyngeal HSV 1 Influenza CNS Mumps virus Rhinovirus JC virus RSV Eye Herpes viruses Parainfluenza HSV Measles Coronavirus Adenovirus LCM virus Cytomegalovirus Flaviviruses Rabies HHV6 Poliovirus Heart Lower respiratory HTLV-1 Coxsackie B virus Rhinoviruses Parainfluenza virus HIV Coronaviruses Respiratory syncytial virus Parainfluenza virus Adenovirus Respiratory syncytial virus Coronaviruses Gastro-intestinal Influenza virus type A and B Human Bocavirus 1 Adenovirus Hepatitis virus type A, B, C, D, E Those that cause -

Diversity and Evolution of Novel Invertebrate DNA Viruses Revealed by Meta-Transcriptomics
viruses Article Diversity and Evolution of Novel Invertebrate DNA Viruses Revealed by Meta-Transcriptomics Ashleigh F. Porter 1, Mang Shi 1, John-Sebastian Eden 1,2 , Yong-Zhen Zhang 3,4 and Edward C. Holmes 1,3,* 1 Marie Bashir Institute for Infectious Diseases and Biosecurity, Charles Perkins Centre, School of Life & Environmental Sciences and Sydney Medical School, The University of Sydney, Sydney, NSW 2006, Australia; [email protected] (A.F.P.); [email protected] (M.S.); [email protected] (J.-S.E.) 2 Centre for Virus Research, Westmead Institute for Medical Research, Westmead, NSW 2145, Australia 3 Shanghai Public Health Clinical Center and School of Public Health, Fudan University, Shanghai 201500, China; [email protected] 4 Department of Zoonosis, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Changping, Beijing 102206, China * Correspondence: [email protected]; Tel.: +61-2-9351-5591 Received: 17 October 2019; Accepted: 23 November 2019; Published: 25 November 2019 Abstract: DNA viruses comprise a wide array of genome structures and infect diverse host species. To date, most studies of DNA viruses have focused on those with the strongest disease associations. Accordingly, there has been a marked lack of sampling of DNA viruses from invertebrates. Bulk RNA sequencing has resulted in the discovery of a myriad of novel RNA viruses, and herein we used this methodology to identify actively transcribing DNA viruses in meta-transcriptomic libraries of diverse invertebrate species. Our analysis revealed high levels of phylogenetic diversity in DNA viruses, including 13 species from the Parvoviridae, Circoviridae, and Genomoviridae families of single-stranded DNA virus families, and six double-stranded DNA virus species from the Nudiviridae, Polyomaviridae, and Herpesviridae, for which few invertebrate viruses have been identified to date. -

Virus World As an Evolutionary Network of Viruses and Capsidless Selfish Elements
Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements Koonin, E. V., & Dolja, V. V. (2014). Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements. Microbiology and Molecular Biology Reviews, 78(2), 278-303. doi:10.1128/MMBR.00049-13 10.1128/MMBR.00049-13 American Society for Microbiology Version of Record http://cdss.library.oregonstate.edu/sa-termsofuse Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements Eugene V. Koonin,a Valerian V. Doljab National Center for Biotechnology Information, National Library of Medicine, Bethesda, Maryland, USAa; Department of Botany and Plant Pathology and Center for Genome Research and Biocomputing, Oregon State University, Corvallis, Oregon, USAb Downloaded from SUMMARY ..................................................................................................................................................278 INTRODUCTION ............................................................................................................................................278 PREVALENCE OF REPLICATION SYSTEM COMPONENTS COMPARED TO CAPSID PROTEINS AMONG VIRUS HALLMARK GENES.......................279 CLASSIFICATION OF VIRUSES BY REPLICATION-EXPRESSION STRATEGY: TYPICAL VIRUSES AND CAPSIDLESS FORMS ................................279 EVOLUTIONARY RELATIONSHIPS BETWEEN VIRUSES AND CAPSIDLESS VIRUS-LIKE GENETIC ELEMENTS ..............................................280 Capsidless Derivatives of Positive-Strand RNA Viruses....................................................................................................280 -

The Intestinal Virome of Malabsorption Syndrome-Affected and Unaffected
Virus Research 261 (2019) 9–20 Contents lists available at ScienceDirect Virus Research journal homepage: www.elsevier.com/locate/virusres The intestinal virome of malabsorption syndrome-affected and unaffected broilers through shotgun metagenomics T ⁎ Diane A. Limaa, , Samuel P. Cibulskib, Caroline Tochettoa, Ana Paula M. Varelaa, Fabrine Finklera, Thais F. Teixeiraa, Márcia R. Loikoa, Cristine Cervac, Dennis M. Junqueirad, Fabiana Q. Mayerc, Paulo M. Roehea a Laboratório de Virologia, Departamento de Microbiologia, Imunologia e Parasitologia, Instituto de Ciências Básicas da Saúde (ICBS), Universidade Federal do Rio Grande do Sul (UFRGS), Porto Alegre, RS, Brazil b Laboratório de Virologia, Faculdade de Veterinária, Universidade Federal do Rio Grande do Sul, Porto Alegre, RS, Brazil c Laboratório de Biologia Molecular, Instituto de Pesquisas Veterinárias Desidério Finamor (IPVDF), Eldorado do Sul, RS, Brazil d Centro Universitário Ritter dos Reis - UniRitter, Health Science Department, Porto Alegre, RS, Brazil ARTICLE INFO ABSTRACT Keywords: Malabsorption syndrome (MAS) is an economically important disease of young, commercially reared broilers, Enteric disorders characterized by growth retardation, defective feather development and diarrheic faeces. Several viruses have Virome been tentatively associated to such syndrome. Here, in order to examine potential associations between enteric Broiler chickens viruses and MAS, the faecal viromes of 70 stool samples collected from diseased (n = 35) and healthy (n = 35) High-throughput sequencing chickens from seven flocks were characterized and compared. Following high-throughput sequencing, a total of 8,347,319 paired end reads, with an average of 231 nt, were generated. Through analysis of de novo assembled contigs, 144 contigs > 1000 nt were identified with hits to eukaryotic viral sequences, as determined by GenBank database. -

The Virome Hunters Diversity Was—And Still Is to This Day— Mostly Uncharacterized
NEWS FEATURE Virus images: colematt / iStock Getty Images Plus The virome hunters diversity was—and still is to this day— mostly uncharacterized. Scientists have since Ambitious efforts to catalog viruses across the globe may facilitate expanded their analyses into other many environments, as well as animal and human our understanding of viral communities and ecology, boost viromes. Indeed, a pure metagenomic analysis infectious disease diagnostics and surveillance, and spur new of human fecal samples revealed a previously unknown virus that represents a large part therapeutics. Charles Schmidt investigates. of the dark matter—as much as 90%—of the human gut virome. Dubbed the crAssphage by Robert Edwards and collaborators from In July, scientists from UC Davis and Columbia and our questions about the viral world are San Diego State because it was pieced together University announced they had isolated a new profound,” says Edward Holmes, a virologist by tool they invented called cross assembly species of the Ebola virus from bats roosting and professor at the University of Sydney in analysis (although its origin in stool seems to inside houses in Sierra Leone. Dubbed Bombali Australia. Along with new species, investigators have been in the minds of the researchers), it after the district where the bats were captured, are turning up vast stretches of what they call was called “one of the most striking feats of this new species is the first Ebola virus to have dark matter—viral sequences unlike any seen metagenomics at that time” by Eugene Koonin its initial identification in an animal host previously. They’re using sophisticated bioin- at US National Center for Biotechnology rather than from a sick person. -

Single Stranded DNA Viruses Associated with Capybara Faeces Sampled in Brazil
viruses Article Single Stranded DNA Viruses Associated with Capybara Faeces Sampled in Brazil Rafaela S. Fontenele 1,2, Cristiano Lacorte 2, Natalia S. Lamas 2, Kara Schmidlin 1, Arvind Varsani 1,3,* and Simone G. Ribeiro 2,* 1 The Biodesign Center for Fundamental and Applied Microbiomics, Center for Evolution and Medicine School of Life Sciences, Arizona State University, Tempe, AZ 85287, USA 2 Embrapa Recursos Genéticos e Biotecnologia, Brasília, DF 70770-017, Brazil 3 Structural Biology Research Unit, Department of Clinical Laboratory Sciences, University of Cape Town, Observatory, Cape Town 7925, South Africa * Correspondence: [email protected] (A.V.); [email protected] (S.G.R.); Tel.: +1-480-727-2093 (A.V.); +55-61-3448-4666/3448-4703 (S.G.R.) Received: 14 June 2019; Accepted: 31 July 2019; Published: 2 August 2019 Abstract: Capybaras (Hydrochoerus hydrochaeris), the world’s largest rodents, are distributed throughout South America. These wild herbivores are commonly found near water bodies and are well adapted to rural and urban areas. There is limited information on the viruses circulating through capybaras. This study aimed to expand the knowledge on the viral diversity associated with capybaras by sampling their faeces. Using a viral metagenomics approach, we identified diverse single-stranded DNA viruses in the capybara faeces sampled in the Distrito Federal, Brazil. A total of 148 complete genomes of viruses in the Microviridae family were identified. In addition, 14 genomoviruses (family Genomoviridae), a novel cyclovirus (family Circoviridae), and a smacovirus (family Smacoviridae) were identified. Also, 37 diverse viruses that cannot be assigned to known families and more broadly referred to as unclassified circular replication associated protein encoding single-stranded (CRESS) DNA viruses were identified. -

Insights Into Circovirus Host Range from the Genomic Fossil Record
bioRxiv preprint doi: https://doi.org/10.1101/246777; this version posted March 8, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Insights into circovirus host range from the genomic fossil record. Running title: Circovirus host range Authors: Tristan P.W. Dennis1*, Peter J. Flynn2,3*, William Marciel de Souza1,4, Joshua B. Singer1, Corrie S. Moreau2, Sam J. Wilson1, Robert J. Gifford1 Affiliations: 1 MRC-University of Glasgow Centre for Virus Research, Glasgow, UK 2 Field Museum of Natural History, Department of Science and Education, Chicago, IL 60605, USA 3University of Chicago, Committee on Evolutionary Biology, Chicago IL 60637, USA 4 Virology Research Center, School of Medicine of Ribeirão Preto of University of São Paulo, Ribeirão Preto, Brazil * These authors contributed equally Corresponding author: Robert J. Gifford MRC-University of Glasgow Centre for Virus Research 464 Bearsden Road Glasgow, Scotland, UK E-mail: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/246777; this version posted March 8, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract A diverse range of DNA sequences derived from circoviruses (family Circoviridae) have been identified in samples obtained from humans and domestic animals, often in association with pathological conditions. -

The Discovery, Distribution and Diversity of DNA Viruses
bioRxiv preprint doi: https://doi.org/10.1101/2020.10.16.342956; this version posted March 17, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Title: The discovery, distribution and diversity of DNA viruses associated with Drosophila melanogaster in Europe Running title: DNA viruses of European Drosophila Key Words: DNA virus, Endogenous viral element, Drosophila, Nudivirus, Galbut virus, Filamentous virus, Adintovirus, Densovirus, Bidnavirus Authors: Megan A. Wallace 1,2 [email protected] 0000-0001-5367-420X Kelsey A. Coffman 3 [email protected] 0000-0002-7609-6286 Clément Gilbert 1,4 [email protected] 0000-0002-2131-7467 Sanjana Ravindran 2 [email protected] 0000-0003-0996-0262 Gregory F. Albery 5 [email protected] 0000-0001-6260-2662 Jessica Abbott 1,6 [email protected] 0000-0002-8743-2089 Eliza Argyridou 1,7 [email protected] 0000-0002-6890-4642 Paola Bellosta 1,8,9 [email protected] 0000-0003-1913-5661 Andrea J. Betancourt 1,10 [email protected] 0000-0001-9351-1413 Hervé Colinet 1,11 [email protected] 0000-0002-8806-3107 Katarina Eric 1,12 [email protected] 0000-0002-3456-2576 Amanda Glaser-Schmitt 1,7 [email protected] 0000-0002-1322-1000 Sonja Grath 1,7 [email protected] 0000-0003-3621-736X Mihailo Jelic 1,13 [email protected] 0000-0002-1637-0933 Maaria Kankare 1,14 [email protected] 0000-0003-1541-9050 Iryna Kozeretska 1,15 [email protected] 0000-0002-6485-1408 Volker Loeschcke 1,16 [email protected] 0000-0003-1450-0754 Catherine Montchamp-Moreau 1,4 [email protected] 0000-0002-5044-9709 Lino Ometto 1,17 [email protected] 0000-0002-2679-625X Banu Sebnem Onder 1,18 [email protected] 0000-0002-3003-248X Dorcas J. -

Intestinal Virome Changes Precede Autoimmunity in Type I Diabetes-Susceptible Children,” by Guoyan Zhao, Tommi Vatanen, Lindsay Droit, Arnold Park, Aleksandar D
Correction MEDICAL SCIENCES Correction for “Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children,” by Guoyan Zhao, Tommi Vatanen, Lindsay Droit, Arnold Park, Aleksandar D. Kostic, Tiffany W. Poon, Hera Vlamakis, Heli Siljander, Taina Härkönen, Anu-Maaria Hämäläinen, Aleksandr Peet, Vallo Tillmann, Jorma Ilonen, David Wang, Mikael Knip, Ramnik J. Xavier, and Herbert W. Virgin, which was first published July 10, 2017; 10.1073/pnas.1706359114 (Proc Natl Acad Sci USA 114: E6166–E6175). The authors wish to note the following: “After publication, we discovered that certain patient-related information in the spreadsheets placed online had information that could conceiv- ably be used to identify, or at least narrow down, the identity of children whose fecal samples were studied. The article has been updated online to remove these potential privacy concerns. These changes do not alter the conclusions of the paper.” Published under the PNAS license. Published online November 19, 2018. www.pnas.org/cgi/doi/10.1073/pnas.1817913115 E11426 | PNAS | November 27, 2018 | vol. 115 | no. 48 www.pnas.org Downloaded by guest on September 26, 2021 Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children Guoyan Zhaoa,1, Tommi Vatanenb,c, Lindsay Droita, Arnold Parka, Aleksandar D. Kosticb,2, Tiffany W. Poonb, Hera Vlamakisb, Heli Siljanderd,e, Taina Härkönend,e, Anu-Maaria Hämäläinenf, Aleksandr Peetg,h, Vallo Tillmanng,h, Jorma Iloneni, David Wanga,j, Mikael Knipd,e,k,l, Ramnik J. Xavierb,m, and