Emergence of Previously Unknown Poxviruses
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Munich Outbreak of Cutaneous Cowpox Infection: Transmission by Infected Pet Rats
Acta Derm Venereol 2012; 92: 126–131 INVESTIGATIVE REPORT The Munich Outbreak of Cutaneous Cowpox Infection: Transmission by Infected Pet Rats Sandra VOGEL1, Miklós SÁRDY1, Katharina GLOS2, Hans Christian KOrting1, Thomas RUZICKA1 and Andreas WOLLENBERG1 1Department of Dermatology and Allergology, Ludwig Maximilian University, Munich, and 2Department of Dermatology, Haas and Link Animal Clinic, Germering, Germany Cowpox virus infection of humans is an uncommon, another type of orthopoxvirus, from infected pet prairie potentially fatal, skin disease. It is largely confined to dogs have recently been described in the USA, making Europe, but is not found in Eire, or in the USA, Austral the medical community aware of the risk of transmission asia, or the Middle or Far East. Patients having contact of pox viruses from pets (3). with infected cows, cats, or small rodents sporadically Seven of 8 exposed patients living in the Munich contract the disease from these animals. We report here area contracted cowpox virus infection from an unusual clinical aspects of 8 patients from the Munich area who source: rats infected with cowpox virus bought from had purchased infected pet rats from a local supplier. Pet local pet shops and reputedly from the same supplier rats are a novel potential source of local outbreaks. The caused a clinically distinctive pattern of infection, which morphologically distinctive skin lesions are mostly res was mostly restricted to the patients’ neck and trunk. tricted to the patients’ necks, reflecting the infected ani We report here dermatologically relevant aspects of mals’ contact pattern. Individual lesions vaguely resem our patients in order to alert the medical community to ble orf or Milker’s nodule, but show marked surrounding the possible risk of a zoonotic orthopoxvirus outbreak erythema, firm induration and local adenopathy. -

Guide for Common Viral Diseases of Animals in Louisiana
Sampling and Testing Guide for Common Viral Diseases of Animals in Louisiana Please click on the species of interest: Cattle Deer and Small Ruminants The Louisiana Animal Swine Disease Diagnostic Horses Laboratory Dogs A service unit of the LSU School of Veterinary Medicine Adapted from Murphy, F.A., et al, Veterinary Virology, 3rd ed. Cats Academic Press, 1999. Compiled by Rob Poston Multi-species: Rabiesvirus DCN LADDL Guide for Common Viral Diseases v. B2 1 Cattle Please click on the principle system involvement Generalized viral diseases Respiratory viral diseases Enteric viral diseases Reproductive/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 2 Deer and Small Ruminants Please click on the principle system involvement Generalized viral disease Respiratory viral disease Enteric viral diseases Reproductive/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 3 Swine Please click on the principle system involvement Generalized viral diseases Respiratory viral diseases Enteric viral diseases Reproductive/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 4 Horses Please click on the principle system involvement Generalized viral diseases Neurological viral diseases Respiratory viral diseases Enteric viral diseases Abortifacient/neonatal viral diseases Viral infections affecting the skin Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. B2 5 Dogs Please click on the principle system involvement Generalized viral diseases Respiratory viral diseases Enteric viral diseases Reproductive/neonatal viral diseases Back to the Beginning DCN LADDL Guide for Common Viral Diseases v. -

Comparative Analysis, Distribution, and Characterization of Microsatellites in Orf Virus Genome
www.nature.com/scientificreports OPEN Comparative analysis, distribution, and characterization of microsatellites in Orf virus genome Basanta Pravas Sahu1, Prativa Majee 1, Ravi Raj Singh1, Anjan Sahoo2 & Debasis Nayak 1* Genome-wide in-silico identifcation of microsatellites or simple sequence repeats (SSRs) in the Orf virus (ORFV), the causative agent of contagious ecthyma has been carried out to investigate the type, distribution and its potential role in the genome evolution. We have investigated eleven ORFV strains, which resulted in the presence of 1,036–1,181 microsatellites per strain. The further screening revealed the presence of 83–107 compound SSRs (cSSRs) per genome. Our analysis indicates the dinucleotide (76.9%) repeats to be the most abundant, followed by trinucleotide (17.7%), mononucleotide (4.9%), tetranucleotide (0.4%) and hexanucleotide (0.2%) repeats. The Relative Abundance (RA) and Relative Density (RD) of these SSRs varied between 7.6–8.4 and 53.0–59.5 bp/ kb, respectively. While in the case of cSSRs, the RA and RD ranged from 0.6–0.8 and 12.1–17.0 bp/kb, respectively. Regression analysis of all parameters like the incident of SSRs, RA, and RD signifcantly correlated with the GC content. But in a case of genome size, except incident SSRs, all other parameters were non-signifcantly correlated. Nearly all cSSRs were composed of two microsatellites, which showed no biasedness to a particular motif. Motif duplication pattern, such as, (C)-x-(C), (TG)- x-(TG), (AT)-x-(AT), (TC)- x-(TC) and self-complementary motifs, such as (GC)-x-(CG), (TC)-x-(AG), (GT)-x-(CA) and (TC)-x-(AG) were observed in the cSSRs. -

Where Do We Stand After Decades of Studying Human Cytomegalovirus?
microorganisms Review Where do we Stand after Decades of Studying Human Cytomegalovirus? 1, 2, 1 1 Francesca Gugliesi y, Alessandra Coscia y, Gloria Griffante , Ganna Galitska , Selina Pasquero 1, Camilla Albano 1 and Matteo Biolatti 1,* 1 Laboratory of Pathogenesis of Viral Infections, Department of Public Health and Pediatric Sciences, University of Turin, 10126 Turin, Italy; [email protected] (F.G.); gloria.griff[email protected] (G.G.); [email protected] (G.G.); [email protected] (S.P.); [email protected] (C.A.) 2 Complex Structure Neonatology Unit, Department of Public Health and Pediatric Sciences, University of Turin, 10126 Turin, Italy; [email protected] * Correspondence: [email protected] These authors contributed equally to this work. y Received: 19 March 2020; Accepted: 5 May 2020; Published: 8 May 2020 Abstract: Human cytomegalovirus (HCMV), a linear double-stranded DNA betaherpesvirus belonging to the family of Herpesviridae, is characterized by widespread seroprevalence, ranging between 56% and 94%, strictly dependent on the socioeconomic background of the country being considered. Typically, HCMV causes asymptomatic infection in the immunocompetent population, while in immunocompromised individuals or when transmitted vertically from the mother to the fetus it leads to systemic disease with severe complications and high mortality rate. Following primary infection, HCMV establishes a state of latency primarily in myeloid cells, from which it can be reactivated by various inflammatory stimuli. Several studies have shown that HCMV, despite being a DNA virus, is highly prone to genetic variability that strongly influences its replication and dissemination rates as well as cellular tropism. In this scenario, the few currently available drugs for the treatment of HCMV infections are characterized by high toxicity, poor oral bioavailability, and emerging resistance. -

Modulation of NF-Κb Signalling by Microbial Pathogens
REVIEWS Modulation of NF‑κB signalling by microbial pathogens Masmudur M. Rahman and Grant McFadden Abstract | The nuclear factor-κB (NF‑κB) family of transcription factors plays a central part in the host response to infection by microbial pathogens, by orchestrating the innate and acquired host immune responses. The NF‑κB proteins are activated by diverse signalling pathways that originate from many different cellular receptors and sensors. Many successful pathogens have acquired sophisticated mechanisms to regulate the NF‑κB signalling pathways by deploying subversive proteins or hijacking the host signalling molecules. Here, we describe the mechanisms by which viruses and bacteria micromanage the host NF‑κB signalling circuitry to favour the continued survival of the pathogen. The nuclear factor-κB (NF-κB) family of transcription Signalling targets upstream of NF‑κB factors regulates the expression of hundreds of genes that NF-κB proteins are tightly regulated in both the cyto- are associated with diverse cellular processes, such as pro- plasm and the nucleus6. Under normal physiological liferation, differentiation and death, as well as innate and conditions, NF‑κB complexes remain inactive in the adaptive immune responses. The mammalian NF‑κB cytoplasm through a direct interaction with proteins proteins are members of the Rel domain-containing pro- of the inhibitor of NF-κB (IκB) family, including IκBα, tein family: RELA (also known as p65), RELB, c‑REL, IκBβ and IκBε (also known as NF-κBIα, NF-κBIβ and the NF-κB p105 subunit (also known as NF‑κB1; which NF-κBIε, respectively); IκB proteins mask the nuclear is cleaved into the p50 subunit) and the NF-κB p100 localization domains in the NF‑κB complex, thus subunit (also known as NF‑κB2; which is cleaved into retaining the transcription complex in the cytoplasm. -

Characterization of Host Micrornas That Respond to DNA Virus Infection in a Crustacean Tianzhi Huang, Dandan Xu and Xiaobo Zhang*
Huang et al. BMC Genomics 2012, 13:159 http://www.biomedcentral.com/1471-2164/13/159 RESEARCH ARTICLE Open Access Characterization of host microRNAs that respond to DNA virus infection in a crustacean Tianzhi Huang, Dandan Xu and Xiaobo Zhang* Abstract Background: MicroRNAs (miRNAs) are key posttranscriptional regulators of gene expression that are implicated in many processes of eukaryotic cells. It is known that the expression profiles of host miRNAs can be reshaped by viruses. However, a systematic investigation of marine invertebrate miRNAs that respond to virus infection has not yet been performed. Results: In this study, the shrimp Marsupenaeus japonicus was challenged by white spot syndrome virus (WSSV). Small RNA sequencing of WSSV-infected shrimp at different time post-infection (0, 6, 24 and 48 h) identified 63 host miRNAs, 48 of which were conserved in other animals, representing 43 distinct families. Of the identified host miRNAs, 31 were differentially expressed in response to virus infection, of which 25 were up-regulated and six down-regulated. The results were confirmed by northern blots. The TargetScan and miRanda algorithms showed that most target genes of the differentially expressed miRNAs were related to immune responses. Gene ontology analysis revealed that immune signaling pathways were mediated by these miRNAs. Evolutionary analysis showed that three of them, miR-1, miR-7 and miR-34, are highly conserved in shrimp, fruit fly and humans and function in the similar pathways. Conclusions: Our study provides the first large-scale characterization of marine invertebrate miRNAs that respond to virus infection. This will help to reveal the molecular events involved in virus-host interactions mediated by miRNAs and their evolution in animals. -

Specimen Type, Collection Methods, and Diagnostic Assays Available For
Specimen type, collection methods, and diagnostic assays available for the detection of poxviruses from human specimens by the Poxvirus and Rabies Branch, Centers for Disease Control and Prevention1. Specimen Orthopoxvirus Parapoxvirus Yatapoxvirus Molluscipoxvirus Specimen type collection method PCR6 Culture EM8 IHC9,10 Serology11 PCR12 EM8 IHC9,10 PCR13 EM8 PCR EM8 Lesion material Fresh or frozen Swab 5 Lesion material [dry or in media ] [vesicle / pustule Formalin fixed skin, scab / crust, etc.] Paraffin block Fixed slide(s) Container Lesion fluid Swab [vesicle / pustule [dry or in media5] fluid, etc.] Touch prep slide Blood EDTA2 EDTA tube 7 Spun or aliquoted Serum before shipment Spun or aliquoted Plasma before shipment CSF3,4 Sterile 1. The detection of poxviruses by electron microscopy (EM) and immunohistochemical staining (IHC) is performed by the Infectious Disease Pathology Branch of the CDC. 2. EDTA — Ethylenediaminetetraacetic acid. 3. CSF — Cerebrospinal fluid. 4. In order to accurately interpret test results generated from CSF specimens, paired serum must also be submitted. 5. If media is used to store and transport specimens a minimal amount should be used to ensure as little dilution of DNA as possible. 6. Orthopoxvirus generic real-time polymerase chain reaction (PCR) assays will amplify DNA from numerous species of virus within the Orthopoxvirus genus. Species-specific real-time PCR assays are available for selective detection of DNA from variola virus, vaccinia virus, monkeypox virus, and cowpox virus. 7. Blood is not ideal for the detection of orthopoxviruses by PCR as the period of viremia has often passed before sampling occurs. 8. EM can reveal the presence of a poxvirus in clinical specimens or from virus culture, but this technique cannot differentiate between virus species within the same genus. -
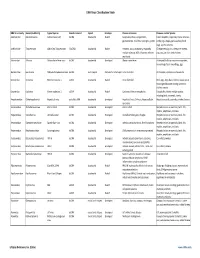
Virus Classification Tables V2.Vd.Xlsx
DNA Virus Classification Table DNA Virus Family Genera (Subfamily) Typical Species Genetic material Capsid Envelope Disease in Humans Diseases in other Species Adenoviridae Mastadenovirus Adenoviruses 1‐47 dsDNA Icosahedral Naked Respiratory illness; conjunctivitis, Canine hepatitis, respiratory illness in horses, gastroenteritis, tonsillitis, meningitis, cystitis cattle, pigs, sheep, goats, sea lions, birds dogs, squirrel enteritis Anelloviridae Torqueviruses Alpha‐Zeta Torqueviruses (‐)ssDNA Icosahedral Naked Hepatitis, lupus, pulmonary, myopathy, Chimpanzee, pig, cow, sheep, tree shrews, multiple sclerosis; 90% of humans infected pigs, cats, sea lions and chickens worldwide Asfarviridae Asfivirus African Swine fever virus dsDNA Icosahedral Enveloped African swine fever Arthropod (tick) transmission or ingestion; hemorrhagic fever in warthogs, pigs Baculoviridae Baculovirus Alpha‐Gamma Baculoviruses dsDNA Stick shaped Occluded or Enveloped none identified Arthropods, Lepidoptera, crustaceans Circoviridae Circovirus Porcine circovirus 1 ssDNA Icosahedral Naked none identified Birds, pigs, dogs; bats; rodents; causes post‐ weaning multisystem wasting syndrome, chicken anemia Circoviridae Cyclovirus Human cyclovirus 1 ssDNA Icosahedral Naked Cyclovirus Vietnam encephalitis Encephalitis; infects multiple species including birds, mammals, insects Hepadnaviridae Orthohepadnavirus Hepatitis B virus partially ssDNA Icosahedral Enveloped Hepatitis B virus; Cirrhosis, Hepatocellular Hepatitis in ducks, squirrels, primates, herons carcinoma Herpesviridae -

Treatment of Metastatic Disease Through Natural Killer Cell Modulation by Infected Cell Vaccines
viruses Review Treatment of Metastatic Disease through Natural Killer Cell Modulation by Infected Cell Vaccines Seyedeh Raheleh Niavarani 1, Christine Lawson 1 and Lee-Hwa Tai 1,2,* 1 Department of Anatomy and Cell Biology, Université de Sherbrooke, Sherbrooke QC J1E 4K8, Canada; [email protected] (S.R.N.); [email protected] (C.L.) 2 Centre de Recherche du Centre Hospitalier de l’Université de Sherbrooke, Sherbrooke QC J1E 4K8, Canada * Correspondence: [email protected]; Tel.: +1-819-821-8000 (ext. 71199) Received: 26 March 2019; Accepted: 10 May 2019; Published: 11 May 2019 Abstract: Oncolytic viruses (OVs) are a form of immunotherapy that release tumor antigens in the context of highly immunogenic viral signals following tumor-targeted infection and destruction. Emerging preclinical and clinical evidence suggests that this in situ vaccine effect is critical for successful viro-immunotherapy. In this review, we discuss the application of OV as an infected cell vaccine (ICV) as one method of enhancing the potency and breadth of anti-tumoral immunity. We focus on understanding and manipulating the critical role of natural killer (NK) cells and their interactions with other immune cells to promote a clinical outcome. With a synergistic tumor killing and immune activating mechanism, ICVs represent a valuable new addition to the cancer fighting toolbox with the potential to treat malignant disease. Keywords: immunotherapy; oncolytic virus; autologous cancer vaccines; infected cell vaccines; natural killer cells; immunomonitoring 1. Introduction While the field of oncology has seen great advances in treating primary solid cancers, malignant cancers that have spread to multiple sites of the body have rarely been cured. -

DNA and Peptide Aptamer Selection for Diagnostic Applications
DNA and Peptide Aptamer Selection for Diagnostic Applications vorgelegt von Diplom-Ingenieurin Janine Michel aus Berlin Von der Fakultät III – Prozesswissenschaften der Technischen Universität Berlin zur Erlangung des akademischen Grades Doktorin der Ingenieurwissenschaften -Dr.-Ing.- genehmigte Dissertation Promotionsausschuss: Vorsitzender: Prof. Dr. Leif-A. Garbe Berichter: Prof. Dr. Jens Kurreck Berichter: PD Dr. Andreas Nitsche Tag der wissenschaftlichen Aussprache: 27.09.2013 Berlin 2013 D83 To Olaf and my loving family, especially to grandpa Bernd. I miss you! I Acknowledgments This work would have been impossible to complete without the help of many persons, including colleagues, family, and friends. Since there are so many of them I cannot acknowledge every single contribution by name, but I would like to thank everyone who helped me through this demanding and challenging but interesting time, regardless of the type of support. Above all, I would like to thank Andreas Nitsche for giving me the opportunity to do my PhD project and for the continuous support. Many thanks go to my dear colleagues Lilija Miller and Daniel Stern who helped me especially in the beginning of this project. Thank you for introducing me to the basics of phage display and aptamers and for the fruitful and valuable scientific discussions and for frequent encouragement. I am grateful to Lilija Miller who helped me with phage display selections and subsequent peptide characterizations. Further, I would like to thank all members of the ZBS1 group for the friendly atmosphere and the company during lunch. A considerable contribution was made by students I supervised during my PhD project. Carolin Ulbricht, Daniel John and Alina Sobiech contributed to the “DNA aptamer selection and characterization project” during their bachelor’s thesis, internships, and master’s thesis, respectively. -

Monkeypox Virus Emergence in Wild Chimpanzees Reveals Distinct Clinical Outcomes and Viral Diversity
ARTICLES https://doi.org/10.1038/s41564-020-0706-0 Monkeypox virus emergence in wild chimpanzees reveals distinct clinical outcomes and viral diversity Livia V. Patrono1,6, Kamilla Pléh1,2,6, Liran Samuni2,3, Markus Ulrich1, Caroline Röthemeier1, Andreas Sachse1, Silvia Muschter4, Andreas Nitsche4, Emmanuel Couacy-Hymann5, Christophe Boesch2,3, Roman M. Wittig 2,3, Sébastien Calvignac-Spencer1 and Fabian H. Leendertz 1 ✉ Monkeypox is a viral zoonotic disease on the rise across endemic habitats. Despite the growing importance of monkeypox virus, our knowledge on its host spectrum and sylvatic maintenance is limited. Here, we describe the recent repeated emergence of monkeypox virus in a wild, human-habituated western chimpanzee (Pan troglodytes verus, hereafter chimpanzee) population from Taï National Park, Ivory Coast. Through daily monitoring, we show that further to causing its typical exanthematous syn- drome, monkeypox can present itself as a severe respiratory disease without a diffuse rash. By analysing 949 non-invasively collected samples, we identify the circulation of at least two distinct monkeypox virus lineages and document the shedding of infectious particles in faeces and flies, suggesting that they could mediate indirect transmission. We also show that the carnivo- rous component of the Taï chimpanzees’ diet, mainly consisting of the sympatric monkeys they regularly hunt, did not change nor shift towards rodent consumption (the presumed reservoir) before the outbreaks, suggesting that the sudden emergence of monkeypox virus in this population is probably due to changes in the ecology of the virus itself. Using long-term mortality surveillance data from Taï National Park, we provide evidence of little to no prior viral activity over at least two decades. -

Effects of Pre-Existing Orthopoxvirus-Specific Immunity On
www.nature.com/scientificreports OPEN Efects of pre-existing orthopoxvirus-specifc immunity on the performance of Modifed Received: 4 January 2018 Accepted: 10 April 2018 Vaccinia virus Ankara-based Published: xx xx xxxx infuenza vaccines Arwen F. Altenburg1, Stella E. van Trierum1, Erwin de Bruin1, Dennis de Meulder1, Carolien E. van de Sandt1, Fiona R. M. van der Klis2, Ron A. M. Fouchier1, Marion P. G. Koopmans1, Guus F. Rimmelzwaan1,3 & Rory D. de Vries1 The replication-defcient orthopoxvirus modifed vaccinia virus Ankara (MVA) is a promising vaccine vector against various pathogens and has an excellent safety record. However, pre-existing vector- specifc immunity is frequently suggested to be a drawback of MVA-based vaccines. To address this issue, mice were vaccinated with MVA-based infuenza vaccines in the presence or absence of orthopoxvirus-specifc immunity. Importantly, protective efcacy of an MVA-based infuenza vaccine against a homologous challenge was not impaired in the presence of orthopoxvirus-specifc pre-existing immunity. Nonetheless, orthopoxvirus-specifc pre-existing immunity reduced the induction of antigen- specifc antibodies under specifc conditions and completely prevented induction of antigen-specifc T cell responses by rMVA-based vaccination. Notably, antibodies induced by vaccinia virus vaccination, both in mice and humans, were not capable of neutralizing MVA. Thus, when using rMVA-based vaccines it is important to consider the main correlate of protection induced by the vaccine, the vaccine dose and the orthopoxvirus immune status of vaccine recipients. Recombinant viral vectors are under development as novel vaccine candidates that induce immunity to antigens of interest expressed from transgenes.