Serine/Threonine Kinase Profiling Using the LANCE Ultra Kinaselect
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Polo-Like Kinases Mediate Cell Survival in Mitochondrial Dysfunction
Polo-like kinases mediate cell survival in mitochondrial dysfunction Takumi Matsumotoa,1, Ping-yuan Wanga,1, Wenzhe Maa, Ho Joong Sunga, Satoaki Matobab, and Paul M. Hwanga,2 aTranslational Medicine Branch, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD 20892; and bCardiovascular Medicine, Kyoto Prefectural University of Medicine, Kyoto 602-8566, Japan Edited by Solomon H. Snyder, Johns Hopkins University School of Medicine, Baltimore, MD, and approved July 10, 2009 (received for review April 16, 2009) Cancer cells often display defects in mitochondrial respiration, thus significant respiration (Fig. S1). For the described in vitro the identification of pathways that promote cell survival under this experiments, one representative SCO2-/- cell line was used. metabolic state may have therapeutic implications. Here, we report However, all significant findings were reproduced or confirmed that the targeted ablation of mitochondrial respiration markedly using at least one additional SCO2-/- cell line that was obtained increases expression of Polo-like kinase 2 (PLK2) and that it is by an independent homologous recombination event to rule out required for the in vitro growth of these nonrespiring cells. clonal variability. Furthermore, we identify PLK2 as a kinase that phosphorylates In an attempt to identify genes associated with the cell cycle Ser-137 of PLK1, which is sufficient to mediate this survival signal. that may enable the survival of SCO2-/- cells after disruption of In vivo, knockdown of PLK2 in an isogenic human cell line with a respiration, we compared microarray gene expression of respir- modest defect in mitochondrial respiration eliminates xenograft ing SCO2ϩ/ϩ and nonrespiring SCO2-/- HCT116 human colon formation, indicating that PLK2 activity is necessary for growth of cancer cells (Table S1). -

The Drug Sensitivity and Resistance Testing (DSRT) Approach
A phenotypic screening and machine learning platform eciently identifies triple negative breast cancer-selective and readily druggable targets Prson Gautam 1 Alok Jaiswal 1 Tero Aittokallio 1, 2 Hassan Al Ali 3 Krister Wennerberg 1,4 Identifying eective oncogenic targets is challenged by the complexity of genetic alterations in 1Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Finland cancer and their poorly understood relation to cell function and survival. There is a need for meth- Current kinome coverage of kinase inhibitors in TNBC exhibit diverse kinase dependencies MFM-223 is selectively addicted to FGFR2 2Department of Mathematics and Statistics, University of Turku, Finland 3The Miami Project to Cure Paralysis, Peggy and Harold Katz Family Drug Discovery Center, A A Sylvester Comprehensive Cancer Center, and Department of Neurological Surgery and Medicine ods that rapidly and accurately identify “pharmacologically eective” targets without the require- clinical evaluation TN Kinases MFM-223 CAL-120 MDA-MB-231 TNBC TNBC TNBC TNBC TNBC TNBC HER2+ 100 University of Miami Miller School of Medicine, Miami, FL 33136, USA. non- HER2+ FGFR1 0.97 0.00 0.00 MFM-223 BL1 BL2 M MSL IM LAR ER+, PR+ 50 ment for priori knowledge of complex signaling networks. We developed an approach that uses ma- cancerous FGFR2 56.46 0.00 0.00 CAL-120 25 4 MDA-MB-231 Biotech Research & Innovation Centre (BRIC) and Novo Nordisk Foundation Center HCC1937 CAL-85-1 CAL-120 MDA-MB-231 DU4475 CAL-148 MCF-10A SK-BR-3 BT-474 FGFR3 25.10 0.00 0.00 0 chine learning to relate results from unbiased phenotypic screening of kinase inhibitors to their bio- for Stem Cell Biology (DanStem), University of Copenhagen, Denmark HCC1599 HDQ-P1 BT-549 MDA-MB-436 MFM-223 FGFR4 0.00 0.00 0.00 MAXIS*Bk Clinical status MDA-MB-468 CAL-51 Hs578T MDA-MB-453 score chemical activity data. -

'Stress–Response' Kinase Pathways in Alzheimer's Disease Progression
Available online at www.sciencedirect.com ScienceDirect Involvement of ‘stress–response’ kinase pathways in Alzheimer’s disease progression 1 Georges Mairet-Coello and Franck Polleux Alzheimer’s disease (AD) is the most prevalent cause of oligomers of Ab are causal to synaptic toxicity [2], trigger dementia, affecting more than 25 million people worldwide. synaptic dysfunction, synapse loss and impaired long- Current models of the pathophysiological mechanisms of AD term potentiation (LTP) [3]. The exact nature of the suggest that the accumulation of soluble oligomeric forms of Ab species (dimers, trimers, Ab*56, protofibrils) respon- amyloid-b (Ab) peptides causes early loss of excitatory sible for this early synaptotoxicity is still under investi- synapses and impairs synaptic plasticity. The signaling gation [4]. Most experimental designs use a mixture of pathways mediating Ab oligomer-induced impairment of various forms of synthetic Ab oligomers, or natural Ab synaptic plasticity and loss of excitatory synapses are only oligomers isolated from the brain of AD subjects, to beginning to be unraveled. Here, we review recent evidence induce rapid loss of excitatory synapses in hippocampal supporting the critical contribution of conserved ‘stress– and cortical neurons in vitro [5–10]. Transgenic mouse response’ kinase pathways in AD progression. models of AD engineered to overexpress human mutant forms of Amyloid Precursor Protein (hAPP), with or with- Addresses out overexpression of mutant presenilins (PS), produce The Scripps Research Institute, Dorris Neuroscience Center, Department of Molecular and Cellular Neuroscience, La Jolla, CA 92037- high levels of Ab1–40 and Ab1–42 peptides and oligo- 1000, USA mers, recapitulate the reduction of excitatory synapses, exhibit neuronal network dysfunction and cognitive def- Corresponding author: Polleux, Franck ([email protected]) 1 icits in spatial learning [6,11]. -

AGC Kinases in Mtor Signaling, in Mike Hall and Fuyuhiko Tamanoi: the Enzymes, Vol
Provided for non-commercial research and educational use only. Not for reproduction, distribution or commercial use. This chapter was originally published in the book, The Enzymes, Vol .27, published by Elsevier, and the attached copy is provided by Elsevier for the author's benefit and for the benefit of the author's institution, for non-commercial research and educational use including without limitation use in instruction at your institution, sending it to specific colleagues who know you, and providing a copy to your institution’s administrator. All other uses, reproduction and distribution, including without limitation commercial reprints, selling or licensing copies or access, or posting on open internet sites, your personal or institution’s website or repository, are prohibited. For exceptions, permission may be sought for such use through Elsevier's permissions site at: http://www.elsevier.com/locate/permissionusematerial From: ESTELA JACINTO, AGC Kinases in mTOR Signaling, In Mike Hall and Fuyuhiko Tamanoi: The Enzymes, Vol. 27, Burlington: Academic Press, 2010, pp.101-128. ISBN: 978-0-12-381539-2, © Copyright 2010 Elsevier Inc, Academic Press. Author's personal copy 7 AGC Kinases in mTOR Signaling ESTELA JACINTO Department of Physiology and Biophysics UMDNJ-Robert Wood Johnson Medical School, Piscataway New Jersey, USA I. Abstract The mammalian target of rapamycin (mTOR), a protein kinase with homology to lipid kinases, orchestrates cellular responses to growth and stress signals. Various extracellular and intracellular inputs to mTOR are known. mTOR processes these inputs as part of two mTOR protein com- plexes, mTORC1 or mTORC2. Surprisingly, despite the many cellular functions that are linked to mTOR, there are very few direct mTOR substrates identified to date. -

Advancing a Clinically Relevant Perspective of the Clonal Nature of Cancer
Advancing a clinically relevant perspective of the clonal nature of cancer Christian Ruiza,b, Elizabeth Lenkiewicza, Lisa Eversa, Tara Holleya, Alex Robesona, Jeffrey Kieferc, Michael J. Demeurea,d, Michael A. Hollingsworthe, Michael Shenf, Donna Prunkardf, Peter S. Rabinovitchf, Tobias Zellwegerg, Spyro Moussesc, Jeffrey M. Trenta,h, John D. Carpteni, Lukas Bubendorfb, Daniel Von Hoffa,d, and Michael T. Barretta,1 aClinical Translational Research Division, Translational Genomics Research Institute, Scottsdale, AZ 85259; bInstitute for Pathology, University Hospital Basel, University of Basel, 4031 Basel, Switzerland; cGenetic Basis of Human Disease, Translational Genomics Research Institute, Phoenix, AZ 85004; dVirginia G. Piper Cancer Center, Scottsdale Healthcare, Scottsdale, AZ 85258; eEppley Institute for Research in Cancer and Allied Diseases, Nebraska Medical Center, Omaha, NE 68198; fDepartment of Pathology, University of Washington, Seattle, WA 98105; gDivision of Urology, St. Claraspital and University of Basel, 4058 Basel, Switzerland; hVan Andel Research Institute, Grand Rapids, MI 49503; and iIntegrated Cancer Genomics Division, Translational Genomics Research Institute, Phoenix, AZ 85004 Edited* by George F. Vande Woude, Van Andel Research Institute, Grand Rapids, MI, and approved June 10, 2011 (received for review March 11, 2011) Cancers frequently arise as a result of an acquired genomic insta- on the basis of morphology alone (8). Thus, the application of bility and the subsequent clonal evolution of neoplastic cells with purification methods such as laser capture microdissection does variable patterns of genetic aberrations. Thus, the presence and not resolve the complexities of many samples. A second approach behaviors of distinct clonal populations in each patient’s tumor is to passage tumor biopsies in tissue culture or in xenografts (4, 9– may underlie multiple clinical phenotypes in cancers. -

A Calcium- and Calmodulin-Dependent Kinase I␣/ Microtubule Affinity Regulating Kinase 2 Signaling Cascade Mediates Calcium-Dependent Neurite Outgrowth
The Journal of Neuroscience, April 18, 2007 • 27(16):4413–4423 • 4413 Cellular/Molecular A Calcium- and Calmodulin-Dependent Kinase I␣/ Microtubule Affinity Regulating Kinase 2 Signaling Cascade Mediates Calcium-Dependent Neurite Outgrowth Nataliya V. Uboha,1 Marc Flajolet,2 Angus C. Nairn,1,2 and Marina R. Picciotto1 1Department of Psychiatry, Yale University School of Medicine, New Haven, Connecticut 06508, and 2Laboratory of Molecular and Cellular Neuroscience, The Rockefeller University, New York, New York 10021 Calcium is a critical regulator of neuronal differentiation and neurite outgrowth during development, as well as synaptic plasticity in adulthood. Calcium- and calmodulin-dependent kinase I (CaMKI) can regulate neurite outgrowth; however, the signal transduction cascades that lead to its physiological effects have not yet been elucidated. CaMKI␣ was therefore used as bait in a yeast two-hybrid assay and microtubule affinity regulating kinase 2 (MARK2)/Par-1b was identified as an interacting partner of CaMKI in three independent screens. The interaction between CaMKI and MARK2 was confirmed in vitro and in vivo by coimmunoprecipitation. CaMKI binds MARK2 within its kinase domain, but only if it is activated by calcium and calmodulin. Expression of CaMKI and MARK2 in Neuro-2A (N2a) cells and in primary hippocampal neurons promotes neurite outgrowth, an effect dependent on the catalytic activities of these enzymes. In addition, decreasing MARK2 activity blocks the ability of the calcium ionophore ionomycin to promote neurite outgrowth. Finally, CaMKI phosphorylates MARK2 on novel sites within its kinase domain. Mutation of these phosphorylation sites decreases both MARK2 kinase activity and its ability to promote neurite outgrowth. -

Eef2k) Natural Product and Synthetic Small Molecule Inhibitors for Cancer Chemotherapy
International Journal of Molecular Sciences Review Progress in the Development of Eukaryotic Elongation Factor 2 Kinase (eEF2K) Natural Product and Synthetic Small Molecule Inhibitors for Cancer Chemotherapy Bin Zhang 1 , Jiamei Zou 1, Qiting Zhang 2, Ze Wang 1, Ning Wang 2,* , Shan He 1 , Yufen Zhao 2 and C. Benjamin Naman 1,* 1 Li Dak Sum Yip Yio Chin Kenneth Li Marine Biopharmaceutical Research Center, College of Food and Pharmaceutical Sciences, Ningbo University, Ningbo 315800, China; [email protected] (B.Z.); [email protected] (J.Z.); [email protected] (Z.W.); [email protected] (S.H.) 2 Institute of Drug Discovery Technology, Ningbo University, Ningbo 315211, China; [email protected] (Q.Z.); [email protected] (Y.Z.) * Correspondence: [email protected] (N.W.); [email protected] (C.B.N.) Abstract: Eukaryotic elongation factor 2 kinase (eEF2K or Ca2+/calmodulin-dependent protein kinase, CAMKIII) is a new member of an atypical α-kinase family different from conventional protein kinases that is now considered as a potential target for the treatment of cancer. This protein regulates the phosphorylation of eukaryotic elongation factor 2 (eEF2) to restrain activity and inhibit the elongation stage of protein synthesis. Mounting evidence shows that eEF2K regulates the cell cycle, autophagy, apoptosis, angiogenesis, invasion, and metastasis in several types of cancers. The Citation: Zhang, B.; Zou, J.; Zhang, expression of eEF2K promotes survival of cancer cells, and the level of this protein is increased in Q.; Wang, Z.; Wang, N.; He, S.; Zhao, many cancer cells to adapt them to the microenvironment conditions including hypoxia, nutrient Y.; Naman, C.B. -

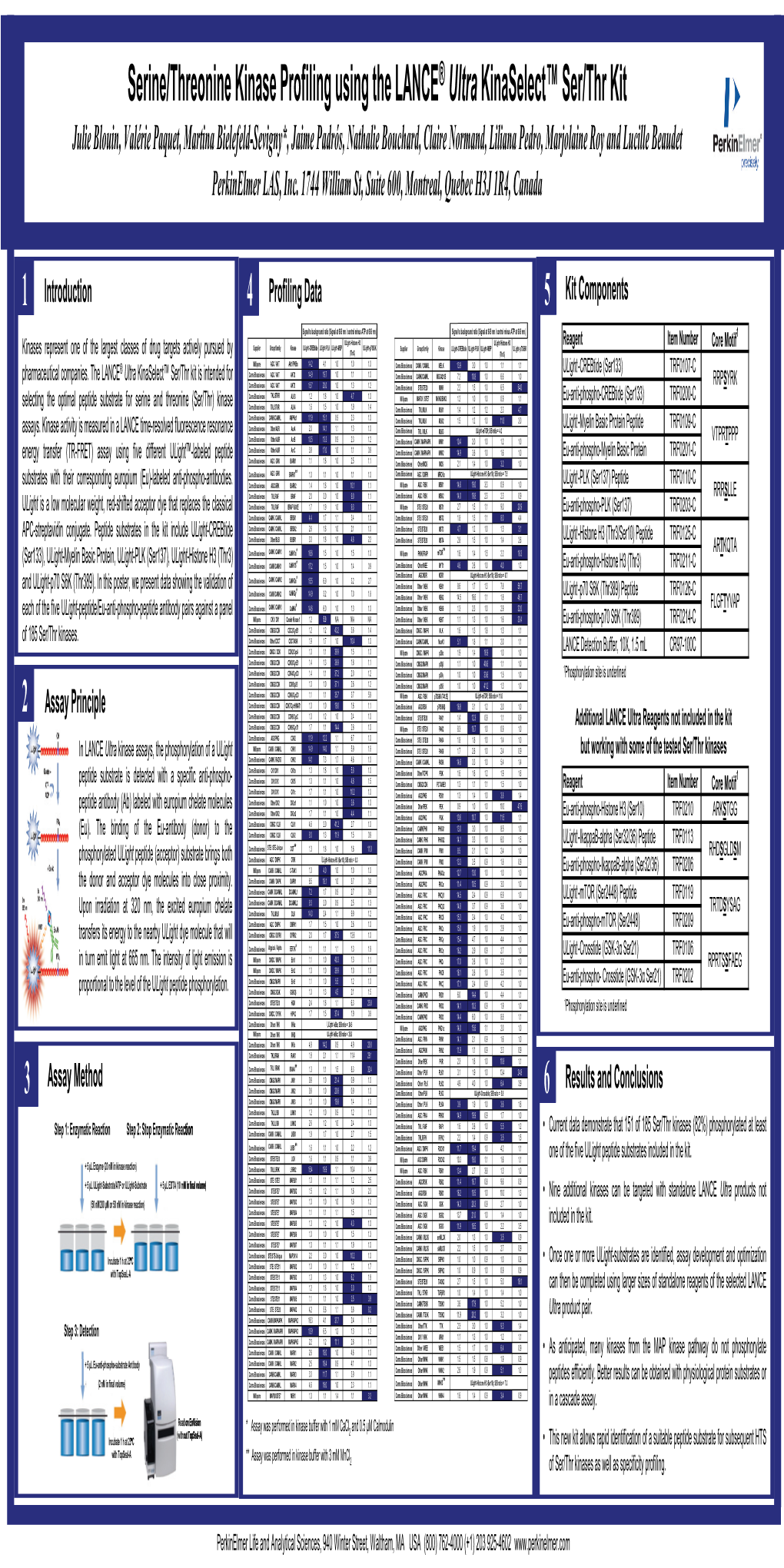
Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

Application of a MYC Degradation
SCIENCE SIGNALING | RESEARCH ARTICLE CANCER Copyright © 2019 The Authors, some rights reserved; Application of a MYC degradation screen identifies exclusive licensee American Association sensitivity to CDK9 inhibitors in KRAS-mutant for the Advancement of Science. No claim pancreatic cancer to original U.S. Devon R. Blake1, Angelina V. Vaseva2, Richard G. Hodge2, McKenzie P. Kline3, Thomas S. K. Gilbert1,4, Government Works Vikas Tyagi5, Daowei Huang5, Gabrielle C. Whiten5, Jacob E. Larson5, Xiaodong Wang2,5, Kenneth H. Pearce5, Laura E. Herring1,4, Lee M. Graves1,2,4, Stephen V. Frye2,5, Michael J. Emanuele1,2, Adrienne D. Cox1,2,6, Channing J. Der1,2* Stabilization of the MYC oncoprotein by KRAS signaling critically promotes the growth of pancreatic ductal adeno- carcinoma (PDAC). Thus, understanding how MYC protein stability is regulated may lead to effective therapies. Here, we used a previously developed, flow cytometry–based assay that screened a library of >800 protein kinase inhibitors and identified compounds that promoted either the stability or degradation of MYC in a KRAS-mutant PDAC cell line. We validated compounds that stabilized or destabilized MYC and then focused on one compound, Downloaded from UNC10112785, that induced the substantial loss of MYC protein in both two-dimensional (2D) and 3D cell cultures. We determined that this compound is a potent CDK9 inhibitor with a previously uncharacterized scaffold, caused MYC loss through both transcriptional and posttranslational mechanisms, and suppresses PDAC anchorage- dependent and anchorage-independent growth. We discovered that CDK9 enhanced MYC protein stability 62 through a previously unknown, KRAS-independent mechanism involving direct phosphorylation of MYC at Ser . -

Supplementary Table 1
SI Table S1. Broad protein kinase selectivity for PF-2771. Kinase, PF-2771 % Inhibition at 10 μM Service Kinase, PF-2771 % Inhibition at 1 μM Service rat RPS6KA1 (RSK1) 39 Dundee AURKA (AURA) 24 Invitrogen IKBKB (IKKb) 26 Dundee CDK2 /CyclinA 21 Invitrogen mouse LCK 25 Dundee rabbit MAP2K1 (MEK1) 19 Dundee AKT1 (AKT) 21 Dundee IKBKB (IKKb) 16 Dundee CAMK1 (CaMK1a) 19 Dundee PKN2 (PRK2) 14 Dundee RPS6KA5 (MSK1) 18 Dundee MAPKAPK5 14 Dundee PRKD1 (PKD1) 13 Dundee PIM3 12 Dundee MKNK2 (MNK2) 12 Dundee PRKD1 (PKD1) 12 Dundee MARK3 10 Dundee NTRK1 (TRKA) 12 Invitrogen SRPK1 9 Dundee MAPK12 (p38g) 11 Dundee MAPKAPK5 9 Dundee MAPK8 (JNK1a) 11 Dundee MAPK13 (p38d) 8 Dundee rat PRKAA2 (AMPKa2) 11 Dundee AURKB (AURB) 5 Dundee NEK2 11 Invitrogen CSK 5 Dundee CHEK2 (CHK2) 11 Invitrogen EEF2K (EEF-2 kinase) 4 Dundee MAPK9 (JNK2) 9 Dundee PRKCA (PKCa) 4 Dundee rat RPS6KA1 (RSK1) 8 Dundee rat PRKAA2 (AMPKa2) 4 Dundee DYRK2 7 Dundee rat CSNK1D (CKId) 3 Dundee AKT1 (AKT) 7 Dundee LYN 3 BioPrint PIM2 7 Invitrogen CSNK2A1 (CKIIa) 3 Dundee MAPK15 (ERK7) 6 Dundee CAMKK2 (CAMKKB) 1 Dundee mouse LCK 5 Dundee PIM3 1 Dundee PDPK1 (PDK1) (directed 5 Invitrogen rat DYRK1A (MNB) 1 Dundee RPS6KB1 (p70S6K) 5 Dundee PBK 0 Dundee CSNK2A1 (CKIIa) 4 Dundee PIM1 -1 Dundee CAMKK2 (CAMKKB) 4 Dundee DYRK2 -2 Dundee SRC 4 Invitrogen MAPK12 (p38g) -2 Dundee MYLK2 (MLCK_sk) 3 Invitrogen NEK6 -3 Dundee MKNK2 (MNK2) 2 Dundee RPS6KB1 (p70S6K) -3 Dundee SRPK1 2 Dundee AKT2 -3 Dundee MKNK1 (MNK1) 2 Dundee RPS6KA3 (RSK2) -3 Dundee CHEK1 (CHK1) 2 Invitrogen rabbit MAP2K1 (MEK1) -4 Dundee