The Drug Sensitivity and Resistance Testing (DSRT) Approach
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Core Lab Brochure
CHOOSE THE MOST TRUSTED LONG-READ TECHNOLOGY FOR YOUR CORE Sequence with Confidence The Sequel® II and IIe Systems are powered by Single Molecule, Real-Time (SMRT®) Sequencing, a technology proven to produce highly accurate long reads, known as HiFi reads, for sequencing data you and your customers can trust. SMRT SEQUENCING IS SMART BUSINESS HiFi Reads: PacBio is the only sequencing technology to offer highly accurate long reads. Because HiFi reads are extremely accurate, downstream analysis is simplified and streamlined, requiring less compute time than the error-prone long reads of other technologies. High Throughput: The Sequel II and IIe Systems have high data yields on robust, highly automated platforms to increase productivity and reduce project costs. Efficient and Easy-To-Use Workflows: Our end-to-end solutions feature library preparation in <3 hours and many push- button analysis workflows, so you can run projects quickly and easily. Support: All of our products are backed by a global team of scientists, bioinformaticians, and engineers who stand ready to provide you with outstanding service. OUTSTANDING PERFORMANCE AND RELIABILITY 99% of runs on the Sequel II System completed successfully Sequel II Systems provide reliable performance with the total bases produced by the PacBio fleet steadily increasing, and 99% of runs completed successfully. “In our experience, the Sequel II System was essentially production-ready right out of the box. We have used it for a range of applications and sample types — from human genome sequencing to metagenome and microbiome profiling to non-model plant and animal genomes — and results have been very good.” — Luke Tallon, Director of the Genomics Resource Center at Maryland Genomics pacb.com/Sequel SMRT SEQUENCING APPLICATIONS – EFFICIENT AND COST EFFECTIVE The Sequel II and IIe Systems support a wide range of applications, each adding unique value to a sequencing study. -

New Contributions in Undergraduate Research
PSU McNair Scholars Online Journal Volume 11 Issue 1 Without Borders: Original Contributions Article 6 in Undergraduate Research 2017 Wings Outstretched: New Contributions in Undergraduate Research Follow this and additional works at: https://pdxscholar.library.pdx.edu/mcnair Let us know how access to this document benefits ou.y Recommended Citation (2017) "Wings Outstretched: New Contributions in Undergraduate Research," PSU McNair Scholars Online Journal: Vol. 11: Iss. 1, Article 6. https://doi.org/10.15760/mcnair.2017.01 This open access Full Issue is distributed under the terms of the Creative Commons Attribution-NonCommercial- ShareAlike 4.0 International License (CC BY-NC-SA 4.0). All documents in PDXScholar should meet accessibility standards. If we can make this document more accessible to you, contact our team. Portland State University McNair Research Journal 2017 Without Borders: Original Contributions in Undergraduate Research 2017 Ronald E. McNair Scholars Journal Portland State University 1 About the Program The Portland State University (PSU) Ronald E. McNair Scholars Program at Portland State University works with motivated and talented undergraduates who want to pursue PhDs. It introduces juniors and seniors who are first-generation and low income, and/or members of under-represented groups to academic research and to effective strategies for getting into and graduating from PhD programs. The McNair Scholars Program has academic-year activities and a full-time summer research internship. Scholars take academic and skills-building seminars and workshops during the year, and each scholar works closely with a faculty mentor on original research in the summer. Scholars present their research findings at the McNair Summer Symposium and at other conferences, and are encouraged to publish their papers in the McNair Journal and other scholarly publications. -

Clinical Utility of Recently Identified Diagnostic, Prognostic, And
Modern Pathology (2017) 30, 1338–1366 1338 © 2017 USCAP, Inc All rights reserved 0893-3952/17 $32.00 Clinical utility of recently identified diagnostic, prognostic, and predictive molecular biomarkers in mature B-cell neoplasms Arantza Onaindia1, L Jeffrey Medeiros2 and Keyur P Patel2 1Instituto de Investigacion Marques de Valdecilla (IDIVAL)/Hospital Universitario Marques de Valdecilla, Santander, Spain and 2Department of Hematopathology, MD Anderson Cancer Center, Houston, TX, USA Genomic profiling studies have provided new insights into the pathogenesis of mature B-cell neoplasms and have identified markers with prognostic impact. Recurrent mutations in tumor-suppressor genes (TP53, BIRC3, ATM), and common signaling pathways, such as the B-cell receptor (CD79A, CD79B, CARD11, TCF3, ID3), Toll- like receptor (MYD88), NOTCH (NOTCH1/2), nuclear factor-κB, and mitogen activated kinase signaling, have been identified in B-cell neoplasms. Chronic lymphocytic leukemia/small lymphocytic lymphoma, diffuse large B-cell lymphoma, follicular lymphoma, mantle cell lymphoma, Burkitt lymphoma, Waldenström macroglobulinemia, hairy cell leukemia, and marginal zone lymphomas of splenic, nodal, and extranodal types represent examples of B-cell neoplasms in which novel molecular biomarkers have been discovered in recent years. In addition, ongoing retrospective correlative and prospective outcome studies have resulted in an enhanced understanding of the clinical utility of novel biomarkers. This progress is reflected in the 2016 update of the World Health Organization classification of lymphoid neoplasms, which lists as many as 41 mature B-cell neoplasms (including provisional categories). Consequently, molecular genetic studies are increasingly being applied for the clinical workup of many of these neoplasms. In this review, we focus on the diagnostic, prognostic, and/or therapeutic utility of molecular biomarkers in mature B-cell neoplasms. -
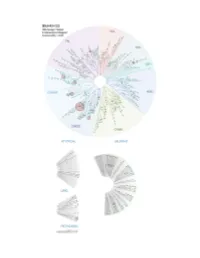
Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) BSJ-03-123 AAK1 AAK1 94 1000 BSJ-03-123 ABL1(E255K)-phosphorylated ABL1 79 1000 BSJ-03-123 ABL1(F317I)-nonphosphorylated ABL1 89 1000 BSJ-03-123 ABL1(F317I)-phosphorylated ABL1 98 1000 BSJ-03-123 ABL1(F317L)-nonphosphorylated ABL1 86 1000 BSJ-03-123 ABL1(F317L)-phosphorylated ABL1 89 1000 BSJ-03-123 ABL1(H396P)-nonphosphorylated ABL1 76 1000 BSJ-03-123 ABL1(H396P)-phosphorylated ABL1 90 1000 BSJ-03-123 ABL1(M351T)-phosphorylated ABL1 100 1000 BSJ-03-123 ABL1(Q252H)-nonphosphorylated ABL1 56 1000 BSJ-03-123 ABL1(Q252H)-phosphorylated ABL1 97 1000 BSJ-03-123 ABL1(T315I)-nonphosphorylated ABL1 100 1000 BSJ-03-123 ABL1(T315I)-phosphorylated ABL1 85 1000 BSJ-03-123 ABL1(Y253F)-phosphorylated ABL1 100 1000 BSJ-03-123 ABL1-nonphosphorylated ABL1 60 1000 BSJ-03-123 ABL1-phosphorylated ABL1 79 1000 BSJ-03-123 ABL2 ABL2 89 1000 BSJ-03-123 ACVR1 ACVR1 100 1000 BSJ-03-123 ACVR1B ACVR1B 95 1000 BSJ-03-123 ACVR2A ACVR2A 100 1000 BSJ-03-123 ACVR2B ACVR2B 96 1000 BSJ-03-123 ACVRL1 ACVRL1 84 1000 BSJ-03-123 ADCK3 CABC1 90 1000 BSJ-03-123 ADCK4 ADCK4 91 1000 BSJ-03-123 AKT1 AKT1 100 1000 BSJ-03-123 AKT2 AKT2 98 1000 BSJ-03-123 AKT3 AKT3 100 1000 BSJ-03-123 ALK ALK 100 1000 BSJ-03-123 ALK(C1156Y) ALK 78 1000 BSJ-03-123 ALK(L1196M) ALK 100 1000 BSJ-03-123 AMPK-alpha1 PRKAA1 93 1000 BSJ-03-123 AMPK-alpha2 PRKAA2 100 1000 BSJ-03-123 ANKK1 ANKK1 89 1000 BSJ-03-123 ARK5 NUAK1 98 1000 BSJ-03-123 ASK1 MAP3K5 100 1000 BSJ-03-123 ASK2 MAP3K6 92 1000 BSJ-03-123 AURKA -

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

Biological Heterogeneity of Chondrosarcoma: from (Epi) Genetics Through Stemness and Deregulated Signaling to Immunophenotype
cancers Review Biological Heterogeneity of Chondrosarcoma: From (Epi) Genetics through Stemness and Deregulated Signaling to Immunophenotype Agnieszka Zaj ˛ac 1 , Sylwia K. Król 2 , Piotr Rutkowski 1 and Anna M. Czarnecka 1,3,* 1 Department of Soft Tissue/Bone Sarcoma and Melanoma, Maria Sklodowska-Curie National Research Institute of Oncology, 02-781 Warsaw, Poland; [email protected] (A.Z.); [email protected] (P.R.) 2 Department of Molecular and Translational Oncology, Maria Sklodowska-Curie National Research Institute of Oncology, 02-781 Warsaw, Poland; [email protected] 3 Department of Experimental Pharmacology, Mossakowski Medical Research Centre, Polish Academy of Sciences, 02-176 Warsaw, Poland * Correspondence: [email protected] Simple Summary: Chondrosarcoma (ChS) is the second most frequently diagnosed malignant bone tumor of cartilaginous origin and is generally resistant to standard treatment options. In this paper, we aim to review the current state of the knowledge regarding ChS. We discuss the genetic, epigenetic, and molecular abnormalities underlying its substantial biological and clinical heterogeneity. This review summarizes the critical genetic and molecular drivers of ChS development and progression, contributing to its radio- and chemotherapy resistance. We describe genomic aberrations and point mutations, as well as epigenetic modifications and deregulated signal transduction pathways. We Citation: Zaj ˛ac,A.; Król, S.K.; provide an insight into the stem-like characteristics and immunophenotype of ChS. The paper also Rutkowski, P.; Czarnecka, A.M. outlines potential diagnostic and prognostic biomarkers of ChS and recently identified novel targets Biological Heterogeneity of for future pharmacological interventions in patients. Chondrosarcoma: From (Epi) Genetics through Stemness and Abstract: Deregulated Signaling to Chondrosarcoma (ChS) is a primary malignant bone tumor. -

Application of a MYC Degradation
SCIENCE SIGNALING | RESEARCH ARTICLE CANCER Copyright © 2019 The Authors, some rights reserved; Application of a MYC degradation screen identifies exclusive licensee American Association sensitivity to CDK9 inhibitors in KRAS-mutant for the Advancement of Science. No claim pancreatic cancer to original U.S. Devon R. Blake1, Angelina V. Vaseva2, Richard G. Hodge2, McKenzie P. Kline3, Thomas S. K. Gilbert1,4, Government Works Vikas Tyagi5, Daowei Huang5, Gabrielle C. Whiten5, Jacob E. Larson5, Xiaodong Wang2,5, Kenneth H. Pearce5, Laura E. Herring1,4, Lee M. Graves1,2,4, Stephen V. Frye2,5, Michael J. Emanuele1,2, Adrienne D. Cox1,2,6, Channing J. Der1,2* Stabilization of the MYC oncoprotein by KRAS signaling critically promotes the growth of pancreatic ductal adeno- carcinoma (PDAC). Thus, understanding how MYC protein stability is regulated may lead to effective therapies. Here, we used a previously developed, flow cytometry–based assay that screened a library of >800 protein kinase inhibitors and identified compounds that promoted either the stability or degradation of MYC in a KRAS-mutant PDAC cell line. We validated compounds that stabilized or destabilized MYC and then focused on one compound, Downloaded from UNC10112785, that induced the substantial loss of MYC protein in both two-dimensional (2D) and 3D cell cultures. We determined that this compound is a potent CDK9 inhibitor with a previously uncharacterized scaffold, caused MYC loss through both transcriptional and posttranslational mechanisms, and suppresses PDAC anchorage- dependent and anchorage-independent growth. We discovered that CDK9 enhanced MYC protein stability 62 through a previously unknown, KRAS-independent mechanism involving direct phosphorylation of MYC at Ser . -

PRKCQ Promotes Oncogenic Growth and Anoikis Resistance of a Subset
Byerly et al. Breast Cancer Research (2016) 18:95 DOI 10.1186/s13058-016-0749-6 RESEARCH ARTICLE Open Access PRKCQ promotes oncogenic growth and anoikis resistance of a subset of triple- negative breast cancer cells Jessica Byerly1†, Gwyneth Halstead-Nussloch1†, Koichi Ito1, Igor Katsyv3 and Hanna Y. Irie1,2* Abstract Background: The protein kinase C (PKC) family comprises distinct classes of proteins, many of which are implicated in diverse cellular functions. Protein tyrosine kinase C theta isoform (PRKCQ)/PKCθ, a member of the novel PKC family, may have a distinct isoform-specific role in breast cancer. PKCθ is preferentially expressed in triple-negative breast cancer (TNBC) compared to other breast tumor subtypes. We hypothesized that PRKCQ/PKCθ critically regulates growth and survival of a subset of TNBC cells. Methods: To elucidate the role of PRKCQ/PKCθ in regulating growth and anoikis resistance, we used both gain and loss of function to modulate expression of PRKCQ. We enhanced expression of PKCθ (kinase-active or inactive) in non-transformed breast epithelial cells (MCF-10A) and assessed effects on epidermal growth factor (EGF)- independent growth, anoikis, and migration. We downregulated expression of PKCθ in TNBC cells, and determined effects on in vitro and in vivo growth and survival. TNBC cells were also treated with a small molecule inhibitor to assess requirement for PKCθ kinase activity in the growth of TNBC cells. Results: PRKCQ/PKCθ can promote oncogenic phenotypes when expressed in non-transformed MCF-10A mammary epithelial cells; PRKCQ/PKCθ enhances anchorage-independent survival, growth-factor-independent proliferation, and migration. PKCθ expression promotes retinoblastoma (Rb) phosphorylation and cell-cycle progression under growth factor-deprived conditions that typically induce cell-cycle arrest of MCF-10A breast epithelial cells. -

Individualized Systems Medicine Strategy to Tailor Treatments for Patients with Chemorefractory Acute Myeloid Leukemia
Published OnlineFirst September 20, 2013; DOI: 10.1158/2159-8290.CD-13-0350 RESEARCH ARTICLE Individualized Systems Medicine Strategy to Tailor Treatments for Patients with Chemorefractory Acute Myeloid Leukemia Tea Pemovska 1 , Mika Kontro 2 , Bhagwan Yadav 1 , Henrik Edgren 1 , Samuli Eldfors1 , Agnieszka Szwajda 1 , Henrikki Almusa 1 , Maxim M. Bespalov 1 , Pekka Ellonen 1 , Erkki Elonen 2 , Bjørn T. Gjertsen5 , 6 , Riikka Karjalainen 1 , Evgeny Kulesskiy 1 , Sonja Lagström 1 , Anna Lehto 1 , Maija Lepistö1 , Tuija Lundán 3 , Muntasir Mamun Majumder 1 , Jesus M. Lopez Marti 1 , Pirkko Mattila 1 , Astrid Murumägi 1 , Satu Mustjoki 2 , Aino Palva 1 , Alun Parsons 1 , Tero Pirttinen 4 , Maria E. Rämet 4 , Minna Suvela 1 , Laura Turunen 1 , Imre Västrik 1 , Maija Wolf 1 , Jonathan Knowles 1 , Tero Aittokallio 1 , Caroline A. Heckman 1 , Kimmo Porkka 2 , Olli Kallioniemi 1 , and Krister Wennerberg 1 ABSTRACT We present an individualized systems medicine (ISM) approach to optimize cancer drug therapies one patient at a time. ISM is based on (i) molecular profi ling and ex vivo drug sensitivity and resistance testing (DSRT) of patients’ cancer cells to 187 oncology drugs, (ii) clinical implementation of therapies predicted to be effective, and (iii) studying consecutive samples from the treated patients to understand the basis of resistance. Here, application of ISM to 28 samples from patients with acute myeloid leukemia (AML) uncovered fi ve major taxonomic drug-response sub- types based on DSRT profi les, some with distinct genomic features (e.g., MLL gene fusions in subgroup IV and FLT3 -ITD mutations in subgroup V). Therapy based on DSRT resulted in several clinical responses. -

( 12 ) United States Patent
US010624968B2 (12 ) United States Patent (10 ) Patent No.: US 10,624,968 B2 Bennett et al. (45 ) Date of Patent : Apr. 21 , 2020 ( 54 ) COMPOUNDS FOR TREATING CANCER WO 2005113554 12/2005 WO 2006078161 7/2006 WO 2006078846 7/2006 (71 ) Applicant: Bicycle Therapeutics Limited , WO 2006122806 11/2006 Cambridge (GB ) WO 2007016176 2/2007 WO 2007044729 4/2007 ( 72 ) Inventors : Gavin Bennett , Cambridge (GB ) ; WO 2007053452 5/2007 Daniel Paul Teufel , Cambridge (GB ) WO 2007070514 6/2007 WO 2007084786 7/2007 WO 2007129161 11/2007 ( 73 ) Assignee : BICYCLERD LIMITED , Cambridge WO 2008039218 4/2008 (GB ) WO 2008109943 9/2008 WO 2008118802 10/2008 ( * ) Notice : Subject to any disclaimer, the term of this WO 2009098450 8/2009 patent is extended or adjusted under 35 WO 2009114512 9/2009 WO 2010089115 8/2010 U.S.C. 154 ( b ) by 0 days . WO 2011090760 7/2011 WO 2013050615 4/2013 (21 ) Appl. No.: 15 /862,964 WO 2016067035 5/2016 WO 2017191460 11/2017 ( 22 ) Filed : Jan. 5 , 2018 WO 2018127699 7/2018 (65 ) Prior Publication Data OTHER PUBLICATIONS US 2018/0200378 A1 Jul. 19 , 2018 Paul Polakis . Antibody Drug Conjugates for Cancer Therapy. Pharmacol Rev. Jan 1 , 2016 ;68 ( 1 ) :3-19 . ( Year: 2016 ) . * Related U.S. Application Data Heinis et al. Phage- encoded combinatorial chemical libraries based (60 ) Provisional application No. 62/ 443,508, filed on Jan. on bicyclic peptides . Nat Chem Biol, 2009 ; 5 ( 7 ) : 502-07. ( Year: 2009) . * 6 , 2017 Eder et al. A phage display derived stabilised bicyclic peptide (51 ) Int. Ci. targeting MMP- 14 shows high imaging contrast in small animal A61K 47/64 ( 2017.01 ) PET imaging. -

( 12 ) Patent Application Publication ( 10 ) Pub . No .: US 2020/0206143 A1
US 20200206143A1 IN (19 ) United States ( 12) Patent Application Publication (10 ) Pub . No.: US 2020/0206143 A1 Moskowitz et al. (43 ) Pub . Date : Jul. 2 , 2020 ( 54 ) PLATELETS LOADED WITH ANTI - CANCER Publication Classification AGENTS (51 ) Int. Ci. A61K 9/50 (2006.01 ) ( 71 ) Applicant: Cellphire, Inc., Rockville , MD (US ) A61K 31/704 (2006.01 ) ( 72 ) Inventors : Keith Andrew Moskowitz , Westfield , A61K 31/502 ( 2006.01 ) IN (US ) ; Rafael Jorda, Bethesda, MD A61K 31/337 (2006.01 ) (US ) ; Ying Yi Zheng , Rockville , MD AOIN 1/02 ( 2006.01) (US ) ; Daniel Sheik , Rockville , MD A61K 9/127 (2006.01 ) (US ) (52 ) U.S. CI. CPC A61K 9/5068 ( 2013.01 ) ; A61K 31/704 (2013.01 ) ; A61K 31/502 ( 2013.01) ; AOIN ( 21 ) Appl. No .: 16 /698,645 1/021 ( 2013.01 ) ; AOIN 1/0226 (2013.01 ) ; A61K 9/1271 (2013.01 ) ; A61K 31/337 ( 22 ) Filed : Nov. 27 , 2019 (2013.01 ) ( 57 ) ABSTRACT Related U.S. Application Data In some embodiments provided herein is a method of (60 ) Provisional application No. 62 / 773,931, filed on Nov. preparing cargo - loaded platelets , comprising : treating plate 30 , 2018 , provisional application No. 62/ 775,141 , lets with a cargo and with a loading buffer comprising a salt , filed on Dec. 4 , 2018, provisional application No. a base , a loading agent , and optionally ethanol, to form the 62 /828,041 , filed on Apr. 2 , 2019 . cargo - loaded platelets . Patent Application Publication Jul. 2 , 2020 Sheet 1 of 20 US 2020/0206143 A1 Amount of Doxorubicin (DOX ) load per CD42b * Platelet DOX(mfi) FIG . 1 Amount of DOX in drug loaded platelets following ADP and /or TRAP stimulation 227 FIG .