Phosphorylation Sites in Protein Kinases and Phosphatases Regulated by Formyl Peptide Receptor 2 Signaling
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

The Role of P21-Activated Protein Kinase 1 in Metabolic Homeostasis
THE ROLE OF P21-ACTIVATED PROTEIN KINASE 1 IN METABOLIC HOMEOSTASIS by YU-TING CHIANG A thesis submitted in conformity with the requirements for the degree of Doctor of Philosophy Graduate Department of Physiology University of Toronto © Copyright by Yu-ting Chiang 2014 The Role of P21-Activated Protein Kinase 1 in Metabolic Homeostasis Yu-ting Chiang Doctor of Philosophy Department of Physiology University of Toronto 2014 Abstract Our laboratory has demonstrated previously that the proglucagon gene (gcg), which encodes the incretin hormone GLP-1, is among the downstream targets of the Wnt signaling pathway; and that Pak1 mediates the stimulatory effect of insulin on Wnt target gene expression in mouse gut non- endocrine cells. Here, I asked whether Pak1 controls gut gcg expression and GLP-1 production, and whether Pak1 deletion leads to impaired metabolic homeostasis in mice. I detected the expression of Pak1 and two other group I Paks in the gut endocrine L cell line GLUTag, and co- localized Pak1 and GLP-1 in the mouse gut. Insulin was shown to stimulate Pak1 Thr423 and β-cat Ser675 phosphorylation. The stimulation of insulin on β-cat Ser675 phosphorylation, gcg promoter activity and gcg mRNA expression could be attenuated by the Pak inhibitor IPA3. Male Pak1-/- mice showed significant reduction in both gut and brain gcg expression levels, and attenuated elevation of plasma GLP-1 levels in response to oral glucose challenge. Notably, the Pak1-/- mice were intolerant to both intraperitoneal and oral glucose administration. Aged Pak1-/- mice showed a severe defect in response to intraperitoneal pyruvate challenge (IPPTT). -

Recombinant MAP2K2 Protein
Recombinant MAP2K2 protein Catalog No: 81332, 81632 Quantity: 20, 1000 µg Expressed In: Baculovirus Concentration: 0.3 µg/µl Source: Human Buffer Contents: Recombinant MAP2K2 protein is supplied in 25 mM HEPES-NaOH pH 7.5, 300 mM NaCl, 10% glycerol, 0.04% Triton X-100, and 0.5 mM TCEP. Background: MAP2K2 (Mitogen-Activated Protein Kinase Kinase Kinase 2) is a dual specificity protein kinase that belongs to the MAP kinase kinase family. This kinase plays a critical role in mitogen growth factor signal transduction. It phosphorylates and activates MAPK1/ERK2 and MAPK2/ERK3. The activation of this kinase itself is dependent on the Ser/Thr phosphorylation by MAP kinase kinase kinases. Mutations in this gene cause cardiofaciocutaneous syndrome (CFC syndrome), a disease characterized by heart defects, cognitive disability, and distinctive facial features similar to those found in Noonan syndrome. Protein Details: Recombinant MAP2K2 protein was expressed in baculovirus expression system as the full length protein (accession number NP_109587.1) with a N-terminal FLAG Tag. The molecular weight of the protein is 45.7 kDa. Application Notes: This product was manufactured as described in Protein Details. Where possible, Active Motif has developed functional or activity assays for Recombinant MAP2K2 protein gel recombinant proteins. Additional characterization such as enzyme kinetic activity 10% SDS-PAGE with Coomassie blue assays, inhibitor screening or other biological activity assays may not have been staining MW: 45.7 kDa performed for every product. All available data for a given product is shown on the lot- Purity: >90% specific Technical Data Sheet. Storage and Guarantee: Recombinant proteins in solution are temperature sensitive and must be stored at -80°C to prevent degradation. -

The Drug Sensitivity and Resistance Testing (DSRT) Approach
A phenotypic screening and machine learning platform eciently identifies triple negative breast cancer-selective and readily druggable targets Prson Gautam 1 Alok Jaiswal 1 Tero Aittokallio 1, 2 Hassan Al Ali 3 Krister Wennerberg 1,4 Identifying eective oncogenic targets is challenged by the complexity of genetic alterations in 1Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Finland cancer and their poorly understood relation to cell function and survival. There is a need for meth- Current kinome coverage of kinase inhibitors in TNBC exhibit diverse kinase dependencies MFM-223 is selectively addicted to FGFR2 2Department of Mathematics and Statistics, University of Turku, Finland 3The Miami Project to Cure Paralysis, Peggy and Harold Katz Family Drug Discovery Center, A A Sylvester Comprehensive Cancer Center, and Department of Neurological Surgery and Medicine ods that rapidly and accurately identify “pharmacologically eective” targets without the require- clinical evaluation TN Kinases MFM-223 CAL-120 MDA-MB-231 TNBC TNBC TNBC TNBC TNBC TNBC HER2+ 100 University of Miami Miller School of Medicine, Miami, FL 33136, USA. non- HER2+ FGFR1 0.97 0.00 0.00 MFM-223 BL1 BL2 M MSL IM LAR ER+, PR+ 50 ment for priori knowledge of complex signaling networks. We developed an approach that uses ma- cancerous FGFR2 56.46 0.00 0.00 CAL-120 25 4 MDA-MB-231 Biotech Research & Innovation Centre (BRIC) and Novo Nordisk Foundation Center HCC1937 CAL-85-1 CAL-120 MDA-MB-231 DU4475 CAL-148 MCF-10A SK-BR-3 BT-474 FGFR3 25.10 0.00 0.00 0 chine learning to relate results from unbiased phenotypic screening of kinase inhibitors to their bio- for Stem Cell Biology (DanStem), University of Copenhagen, Denmark HCC1599 HDQ-P1 BT-549 MDA-MB-436 MFM-223 FGFR4 0.00 0.00 0.00 MAXIS*Bk Clinical status MDA-MB-468 CAL-51 Hs578T MDA-MB-453 score chemical activity data. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Map2k1 and Map2k2 Genes Contribute to the Normal Development of Syncytiotrophoblasts During Placentation
RESEARCH ARTICLE 1363 Development 136, 1363-1374 (2009) doi:10.1242/dev.031872 Map2k1 and Map2k2 genes contribute to the normal development of syncytiotrophoblasts during placentation Valérie Nadeau*, Stéphanie Guillemette*, Louis-François Bélanger, Olivier Jacob, Sophie Roy and Jean Charron† The mammalian genome contains two ERK/MAP kinase kinase genes, Map2k1 and Map2k2, which encode dual-specificity kinases responsible for ERK/MAP kinase activation. In the mouse, loss of Map2k1 function causes embryonic lethality, whereas Map2k2 mutants survive with a normal lifespan, suggesting that Map2k1 masks the phenotype due to the Map2k2 mutation. To uncover the specific function of MAP2K2 and the threshold requirement of MAP2K proteins during embryo formation, we have successively ablated the Map2k gene functions. We report here that Map2k2 haploinsufficiency affects the normal development of placenta in the absence of one Map2k1 allele. Most Map2k1+/–Map2k2+/– embryos die during gestation because of placenta defects restricted to extra-embryonic tissues. The impaired viability of Map2k1+/–Map2k2+/– embryos can be rescued when the Map2k1 deletion is restricted to the embryonic tissues. The severity of the placenta phenotype is dependent on the number of Map2k mutant alleles, the deletion of the Map2k1 allele being more deleterious. Moreover, the deletion of one or both Map2k2 alleles in the context of one null Map2k1 allele leads to the formation of multinucleated trophoblast giant (MTG) cells. Genetic experiments indicate that these structures are derived from Gcm1-expressing syncytiotrophoblasts (SynT), which are affected in their ability to form the uniform SynT layer II lining the maternal sinuses. Thus, even though Map2k1 plays a predominant role, these results enlighten the function of Map2k2 in placenta development. -

Clinical Utility of Recently Identified Diagnostic, Prognostic, And
Modern Pathology (2017) 30, 1338–1366 1338 © 2017 USCAP, Inc All rights reserved 0893-3952/17 $32.00 Clinical utility of recently identified diagnostic, prognostic, and predictive molecular biomarkers in mature B-cell neoplasms Arantza Onaindia1, L Jeffrey Medeiros2 and Keyur P Patel2 1Instituto de Investigacion Marques de Valdecilla (IDIVAL)/Hospital Universitario Marques de Valdecilla, Santander, Spain and 2Department of Hematopathology, MD Anderson Cancer Center, Houston, TX, USA Genomic profiling studies have provided new insights into the pathogenesis of mature B-cell neoplasms and have identified markers with prognostic impact. Recurrent mutations in tumor-suppressor genes (TP53, BIRC3, ATM), and common signaling pathways, such as the B-cell receptor (CD79A, CD79B, CARD11, TCF3, ID3), Toll- like receptor (MYD88), NOTCH (NOTCH1/2), nuclear factor-κB, and mitogen activated kinase signaling, have been identified in B-cell neoplasms. Chronic lymphocytic leukemia/small lymphocytic lymphoma, diffuse large B-cell lymphoma, follicular lymphoma, mantle cell lymphoma, Burkitt lymphoma, Waldenström macroglobulinemia, hairy cell leukemia, and marginal zone lymphomas of splenic, nodal, and extranodal types represent examples of B-cell neoplasms in which novel molecular biomarkers have been discovered in recent years. In addition, ongoing retrospective correlative and prospective outcome studies have resulted in an enhanced understanding of the clinical utility of novel biomarkers. This progress is reflected in the 2016 update of the World Health Organization classification of lymphoid neoplasms, which lists as many as 41 mature B-cell neoplasms (including provisional categories). Consequently, molecular genetic studies are increasingly being applied for the clinical workup of many of these neoplasms. In this review, we focus on the diagnostic, prognostic, and/or therapeutic utility of molecular biomarkers in mature B-cell neoplasms. -
![Viewer 4.0 Software [73]](https://docslib.b-cdn.net/cover/6175/viewer-4-0-software-73-576175.webp)
Viewer 4.0 Software [73]
BMC Genomics BioMed Central Research Open Access Bioinformatic search of plant microtubule-and cell cycle related serine-threonine protein kinases Pavel A Karpov1, Elena S Nadezhdina2,3,AllaIYemets1, Vadym G Matusov1, Alexey Yu Nyporko1,NadezhdaYuShashina3 and Yaroslav B Blume*1 Addresses: 1Institute of Food Biotechnology and Genomics, National Academy of Sciences of Ukraine, 04123 Kyiv, Ukraine, 2Institute of Protein Research, Russian Academy of Sciences, 142290 Pushchino, Moscow Region, Russian Federation and 3AN Belozersky Institute of Physical- Chemical Biology, Moscow State University, Leninsky Gory, 119992 Moscow, Russian Federation E-mail: Pavel A Karpov - [email protected]; Elena S Nadezhdina - [email protected]; Alla I Yemets - [email protected]; Vadym G Matusov - [email protected]; Alexey Yu Nyporko - [email protected]; Nadezhda Yu Shashina - [email protected]; Yaroslav B Blume* - [email protected] *Corresponding author from International Workshop on Computational Systems Biology Approaches to Analysis of Genome Complexity and Regulatory Gene Networks Singapore 20-25 November 2008 Published: 10 February 2010 BMC Genomics 2010, 11(Suppl 1):S14 doi: 10.1186/1471-2164-11-S1-S14 This article is available from: http://www.biomedcentral.com/1471-2164/11/S1/S14 Publication of this supplement was made possible with help from the Bioinformatics Agency for Science, Technology and Research of Singapore and the Institute for Mathematical Sciences at the National University of Singapore. © 2010 Karpov et al; licensee BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. -
Type of the Paper (Article
Supplementary Materials: Identification of Linkages between EDCs in Personal Care Products and Breast Cancer through Data Integration Combined with Gene Network Analysis Hyeri Jeong 1,2, Jongwoon Kim 1,2,* and Youngjun Kim 1,2 Table S1. Interacting genes and their network types of the 27 common related genes between four selected EDCs and ER positive breast cancer based on GeneMANIA network analysis. Degree Centrality Gene Symbol Official Full Name Interacting Gene Networks * AKT1 2 AR 2, 3, 5, 6 BRCA1 2, 3 CASP8 7 EP300 2, 3 ERBB2 2 13 ESR1 Estrogen receptor 1 HDAC5 2 NCOA1 2, 3 NCOA7 2, 3 PIK3CA 2, 3 SLC10A1 1 SMO 1 TP53 2 AKT1 1 AR 2, 7 BCL6 2, 3 BRCA1 2, 3 CASP8 2 EP300 2, 7 12 TP53 Tumor protein p53 ERBB2 1 ESR1 2 HDAC5 2 MTOR 2 NCOA1 2 SMO 1 AKT1 1, 3 AR 2, 3 BRCA1 2 CYP1A1 1 DUSP10 1, 7 Nuclear receptor EP300 1, 2, 3, 6 12 NCOA1 coactivator 1 ESR1 2, 3 HDAC5 7 KLHL24 1 NCOA7 7 PTCH1 7 TP53 2 AR 2 BRCA1 2 EP300 2 ERBB2 1 ESR1 2 AKT serine/threonine 11 AKT1 MAP2K2 1 kinase 1 MTOR 2, 3, 5, 4 NCOA1 1, 3 PIK3CA 2, 3 SMO 3 TP53 1 Int. J. Environ. Res. Public Health 2017, 14 S2 of S7 Table S1. Cont. Degree Centrality Gene Symbol Official Full Name Interacting Gene Networks * ABCG1 7 APOB 1, 7 EP300 2, 7 GABRR1 7 HDAC5 2, 3 11 BCL6 B-cell CLL/lymphoma 6 KLHL24 1 PIK3CA 1 PTCH1 7 SLC10A1 1 SMO 2 TP53 2, 3 AKT1 2 BRCA1 2, 3 CASP8 1, 3, 4 EP300 2 ESR1 2, 3, 5, 6 10 AR Androgen receptor NCOA1 2, 3 PIK3CA 3 SLC10A1 1 SMO 1 TP53 2, 4 AKT1 2 AR 2 BCL6 2, 7 BRCA1 2, 7 E1A binding protein 9 EP300 ESR1 2, 3 p300 KLHL24 1 NCOA1 1, 2, 3, 6 PIK3CA 1 TP53 2, 7 ABCG1 1 AKT1 2, 3 AR 3 Phosphatidylinositol- BCL6 1 4,5-bisphosphate 9 PIK3CA EP300 1 3-kinase catalytic ERBB2 3 subunit alpha ESR1 2, 3 KLHL24 1 MTOR 3, 6 AR 1 AKT1 3 BCL6 2 Smoothened, frizzled CYP1A1 1 8 SMO class receptor ESR1 1 ERBB2 1 PTCH1 2, 3 TP53 1 AKT1 2 AR 2, 3 EP300 2, 7 BRCA1, DNA 7 BRCA1 ESR1 2, 3 repair associated MTOR 2 NCOA1 2 TP53 2, 3 AR 2, 3, 4 CYP1A1 1 ESR1 7 7 CASP8 Caspase 8 GABRA6 7 KLHL24 7 MTOR 7 TP53 2 Int. -
Thejournalofcellb Io Logy
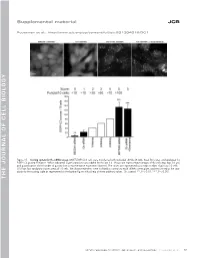
Supplemental material JCB Puustinen et al., http://www.jcb.org/cgi/content/full/jcb.201304012/DC1 Figure S1. Scoring system for the siRNA screen. MCF7-EGFP-LC3 cells were transfected with indicated siRNAs (8 nM), fixed 56 h later, and analyzed for EGFP-LC3 puncta formation. When indicated, 5 µM siramesine was added for the last 3 h. Shown are representative images of the cells (top; bar, 10 µm) and quantification of the number of puncta from a representative experiment (bottom). The values are represented as a mean number of puncta/10 cells ± SDs from four randomly chosen areas of 10 cells. The shown treatments were included as controls to each siRNA screen plate, and they served as the stan- dards for the scoring scale as represented on the bottom figure with a help of three arbitrary values. Ctr, control. **, P < 0.01; ***, P < 0.001. THE JOURNAL OF CELL BIOLOGY CIP2A regulates mTORC1, cell growth, and autophagy • Puustinen et al. S1 Figure S2. A cartoon highlighting the positions of the candidate autophagy-regulating genes in the insulin receptor signaling network. The cartoon was prepared by Ingenuity Pathway Analysis. software. CP, carbohydrate phosphatase; ERK, extracellular signal–regulated kinase; FSH, follicle-stimulating hor- mone; EG, epidermal growth factor. S2 JCB Figure S3. The effect of siRNAs targeting the autophagy-regulating PP2 subunits on target gene expression, cell density, and mTORC1 pathway. (A) Total RNA was isolated from MCF7 cells treated with indicated siRNAs (20 nM) for 54 h and analyzed for target gene expression by qPCR using primers de- signed to specifically amplify the indicated PP2A-related mRNAs andACTB mRNA (internal control). -

Identification of Potential Key Genes and Pathway Linked with Sporadic Creutzfeldt-Jakob Disease Based on Integrated Bioinformatics Analyses
medRxiv preprint doi: https://doi.org/10.1101/2020.12.21.20248688; this version posted December 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Identification of potential key genes and pathway linked with sporadic Creutzfeldt-Jakob disease based on integrated bioinformatics analyses Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India NOTE: This preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. medRxiv preprint doi: https://doi.org/10.1101/2020.12.21.20248688; this version posted December 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. All rights reserved. No reuse allowed without permission. Abstract Sporadic Creutzfeldt-Jakob disease (sCJD) is neurodegenerative disease also called prion disease linked with poor prognosis. The aim of the current study was to illuminate the underlying molecular mechanisms of sCJD. The mRNA microarray dataset GSE124571 was downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) were screened. -

Supplementary Materials
Supplementary materials Supplementary Table S1: MGNC compound library Ingredien Molecule Caco- Mol ID MW AlogP OB (%) BBB DL FASA- HL t Name Name 2 shengdi MOL012254 campesterol 400.8 7.63 37.58 1.34 0.98 0.7 0.21 20.2 shengdi MOL000519 coniferin 314.4 3.16 31.11 0.42 -0.2 0.3 0.27 74.6 beta- shengdi MOL000359 414.8 8.08 36.91 1.32 0.99 0.8 0.23 20.2 sitosterol pachymic shengdi MOL000289 528.9 6.54 33.63 0.1 -0.6 0.8 0 9.27 acid Poricoic acid shengdi MOL000291 484.7 5.64 30.52 -0.08 -0.9 0.8 0 8.67 B Chrysanthem shengdi MOL004492 585 8.24 38.72 0.51 -1 0.6 0.3 17.5 axanthin 20- shengdi MOL011455 Hexadecano 418.6 1.91 32.7 -0.24 -0.4 0.7 0.29 104 ylingenol huanglian MOL001454 berberine 336.4 3.45 36.86 1.24 0.57 0.8 0.19 6.57 huanglian MOL013352 Obacunone 454.6 2.68 43.29 0.01 -0.4 0.8 0.31 -13 huanglian MOL002894 berberrubine 322.4 3.2 35.74 1.07 0.17 0.7 0.24 6.46 huanglian MOL002897 epiberberine 336.4 3.45 43.09 1.17 0.4 0.8 0.19 6.1 huanglian MOL002903 (R)-Canadine 339.4 3.4 55.37 1.04 0.57 0.8 0.2 6.41 huanglian MOL002904 Berlambine 351.4 2.49 36.68 0.97 0.17 0.8 0.28 7.33 Corchorosid huanglian MOL002907 404.6 1.34 105 -0.91 -1.3 0.8 0.29 6.68 e A_qt Magnogrand huanglian MOL000622 266.4 1.18 63.71 0.02 -0.2 0.2 0.3 3.17 iolide huanglian MOL000762 Palmidin A 510.5 4.52 35.36 -0.38 -1.5 0.7 0.39 33.2 huanglian MOL000785 palmatine 352.4 3.65 64.6 1.33 0.37 0.7 0.13 2.25 huanglian MOL000098 quercetin 302.3 1.5 46.43 0.05 -0.8 0.3 0.38 14.4 huanglian MOL001458 coptisine 320.3 3.25 30.67 1.21 0.32 0.9 0.26 9.33 huanglian MOL002668 Worenine