Endometrial Gene Expression in the Early Luteal Phase Is Impacted By
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Viewed Under 23 (B) Or 203 (C) fi M M Male Cko Mice, and Largely Unaffected Magni Cation; Scale Bars, 500 M (B) and 50 M (C)
BRIEF COMMUNICATION www.jasn.org Renal Fanconi Syndrome and Hypophosphatemic Rickets in the Absence of Xenotropic and Polytropic Retroviral Receptor in the Nephron Camille Ansermet,* Matthias B. Moor,* Gabriel Centeno,* Muriel Auberson,* † † ‡ Dorothy Zhang Hu, Roland Baron, Svetlana Nikolaeva,* Barbara Haenzi,* | Natalya Katanaeva,* Ivan Gautschi,* Vladimir Katanaev,*§ Samuel Rotman, Robert Koesters,¶ †† Laurent Schild,* Sylvain Pradervand,** Olivier Bonny,* and Dmitri Firsov* BRIEF COMMUNICATION *Department of Pharmacology and Toxicology and **Genomic Technologies Facility, University of Lausanne, Lausanne, Switzerland; †Department of Oral Medicine, Infection, and Immunity, Harvard School of Dental Medicine, Boston, Massachusetts; ‡Institute of Evolutionary Physiology and Biochemistry, St. Petersburg, Russia; §School of Biomedicine, Far Eastern Federal University, Vladivostok, Russia; |Services of Pathology and ††Nephrology, Department of Medicine, University Hospital of Lausanne, Lausanne, Switzerland; and ¶Université Pierre et Marie Curie, Paris, France ABSTRACT Tight control of extracellular and intracellular inorganic phosphate (Pi) levels is crit- leaves.4 Most recently, Legati et al. have ical to most biochemical and physiologic processes. Urinary Pi is freely filtered at the shown an association between genetic kidney glomerulus and is reabsorbed in the renal tubule by the action of the apical polymorphisms in Xpr1 and primary fa- sodium-dependent phosphate transporters, NaPi-IIa/NaPi-IIc/Pit2. However, the milial brain calcification disorder.5 How- molecular identity of the protein(s) participating in the basolateral Pi efflux remains ever, the role of XPR1 in the maintenance unknown. Evidence has suggested that xenotropic and polytropic retroviral recep- of Pi homeostasis remains unknown. Here, tor 1 (XPR1) might be involved in this process. Here, we show that conditional in- we addressed this issue in mice deficient for activation of Xpr1 in the renal tubule in mice resulted in impaired renal Pi Xpr1 in the nephron. -

The Drug Sensitivity and Resistance Testing (DSRT) Approach
A phenotypic screening and machine learning platform eciently identifies triple negative breast cancer-selective and readily druggable targets Prson Gautam 1 Alok Jaiswal 1 Tero Aittokallio 1, 2 Hassan Al Ali 3 Krister Wennerberg 1,4 Identifying eective oncogenic targets is challenged by the complexity of genetic alterations in 1Institute for Molecular Medicine Finland (FIMM), HiLIFE, University of Helsinki, Finland cancer and their poorly understood relation to cell function and survival. There is a need for meth- Current kinome coverage of kinase inhibitors in TNBC exhibit diverse kinase dependencies MFM-223 is selectively addicted to FGFR2 2Department of Mathematics and Statistics, University of Turku, Finland 3The Miami Project to Cure Paralysis, Peggy and Harold Katz Family Drug Discovery Center, A A Sylvester Comprehensive Cancer Center, and Department of Neurological Surgery and Medicine ods that rapidly and accurately identify “pharmacologically eective” targets without the require- clinical evaluation TN Kinases MFM-223 CAL-120 MDA-MB-231 TNBC TNBC TNBC TNBC TNBC TNBC HER2+ 100 University of Miami Miller School of Medicine, Miami, FL 33136, USA. non- HER2+ FGFR1 0.97 0.00 0.00 MFM-223 BL1 BL2 M MSL IM LAR ER+, PR+ 50 ment for priori knowledge of complex signaling networks. We developed an approach that uses ma- cancerous FGFR2 56.46 0.00 0.00 CAL-120 25 4 MDA-MB-231 Biotech Research & Innovation Centre (BRIC) and Novo Nordisk Foundation Center HCC1937 CAL-85-1 CAL-120 MDA-MB-231 DU4475 CAL-148 MCF-10A SK-BR-3 BT-474 FGFR3 25.10 0.00 0.00 0 chine learning to relate results from unbiased phenotypic screening of kinase inhibitors to their bio- for Stem Cell Biology (DanStem), University of Copenhagen, Denmark HCC1599 HDQ-P1 BT-549 MDA-MB-436 MFM-223 FGFR4 0.00 0.00 0.00 MAXIS*Bk Clinical status MDA-MB-468 CAL-51 Hs578T MDA-MB-453 score chemical activity data. -

Supplementary Data
Supplemental Data A novel mouse model of X-linked nephrogenic diabetes insipidus: Phenotypic analysis and therapeutic implications Jian Hua Li, Chung-Lin Chou, Bo Li, Oksana Gavrilova, Christoph Eisner, Jürgen Schnermann, Stasia A. Anderson, Chu-Xia Deng, Mark A. Knepper, and Jürgen Wess Supplemental Methods Metabolic cage studies. Animals were maintained in mouse metabolic cages (Hatteras Instruments, Cary, NC) under controlled temperature and light conditions (12 hr light and dark cycles). Mice received a fixed daily ration of 6.5 g of gelled diet per 20 g of body weight per day. The gelled diet was composed of 4 g of Basal Diet 5755 (Test Diet, Richmond, IN), 2.5 ml of deionized water, and 65 mg agar. Preweighted drinking water was provided ad libitum during the course of the study. Mice were acclimated in the metabolic cages for 1-2 days. Urine was collected under mineral oil in preweighted collection vials for successive 24 hr periods. Analysis of GPCR expression in mouse IMCD cells via TaqMan real-time qRT-PCR. Total RNA prepared from mouse IMCD tubule suspensions was reverse transcribed as described under Experimental Procedures. Tissues from ten 10-week old C57BL/6 WT mice were collected and pooled for each individual experiment. cDNA derived from 640 ng of RNA was mixed with an equal volume of TaqMan gene expression 2 x master mix (Applied Biosystems, Foster City, CA). 100 μl-aliquots of this mixture (corresponding to 80 ng of RNA) were added to each of the 8 fill ports of a 384-well plate of a mouse GPCR array panel (Applied Biosystems). -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
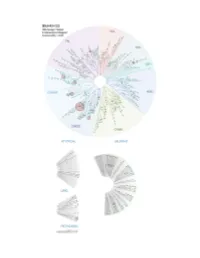
Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) BSJ-03-123 AAK1 AAK1 94 1000 BSJ-03-123 ABL1(E255K)-phosphorylated ABL1 79 1000 BSJ-03-123 ABL1(F317I)-nonphosphorylated ABL1 89 1000 BSJ-03-123 ABL1(F317I)-phosphorylated ABL1 98 1000 BSJ-03-123 ABL1(F317L)-nonphosphorylated ABL1 86 1000 BSJ-03-123 ABL1(F317L)-phosphorylated ABL1 89 1000 BSJ-03-123 ABL1(H396P)-nonphosphorylated ABL1 76 1000 BSJ-03-123 ABL1(H396P)-phosphorylated ABL1 90 1000 BSJ-03-123 ABL1(M351T)-phosphorylated ABL1 100 1000 BSJ-03-123 ABL1(Q252H)-nonphosphorylated ABL1 56 1000 BSJ-03-123 ABL1(Q252H)-phosphorylated ABL1 97 1000 BSJ-03-123 ABL1(T315I)-nonphosphorylated ABL1 100 1000 BSJ-03-123 ABL1(T315I)-phosphorylated ABL1 85 1000 BSJ-03-123 ABL1(Y253F)-phosphorylated ABL1 100 1000 BSJ-03-123 ABL1-nonphosphorylated ABL1 60 1000 BSJ-03-123 ABL1-phosphorylated ABL1 79 1000 BSJ-03-123 ABL2 ABL2 89 1000 BSJ-03-123 ACVR1 ACVR1 100 1000 BSJ-03-123 ACVR1B ACVR1B 95 1000 BSJ-03-123 ACVR2A ACVR2A 100 1000 BSJ-03-123 ACVR2B ACVR2B 96 1000 BSJ-03-123 ACVRL1 ACVRL1 84 1000 BSJ-03-123 ADCK3 CABC1 90 1000 BSJ-03-123 ADCK4 ADCK4 91 1000 BSJ-03-123 AKT1 AKT1 100 1000 BSJ-03-123 AKT2 AKT2 98 1000 BSJ-03-123 AKT3 AKT3 100 1000 BSJ-03-123 ALK ALK 100 1000 BSJ-03-123 ALK(C1156Y) ALK 78 1000 BSJ-03-123 ALK(L1196M) ALK 100 1000 BSJ-03-123 AMPK-alpha1 PRKAA1 93 1000 BSJ-03-123 AMPK-alpha2 PRKAA2 100 1000 BSJ-03-123 ANKK1 ANKK1 89 1000 BSJ-03-123 ARK5 NUAK1 98 1000 BSJ-03-123 ASK1 MAP3K5 100 1000 BSJ-03-123 ASK2 MAP3K6 92 1000 BSJ-03-123 AURKA -

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

PRKCQ Promotes Oncogenic Growth and Anoikis Resistance of a Subset
Byerly et al. Breast Cancer Research (2016) 18:95 DOI 10.1186/s13058-016-0749-6 RESEARCH ARTICLE Open Access PRKCQ promotes oncogenic growth and anoikis resistance of a subset of triple- negative breast cancer cells Jessica Byerly1†, Gwyneth Halstead-Nussloch1†, Koichi Ito1, Igor Katsyv3 and Hanna Y. Irie1,2* Abstract Background: The protein kinase C (PKC) family comprises distinct classes of proteins, many of which are implicated in diverse cellular functions. Protein tyrosine kinase C theta isoform (PRKCQ)/PKCθ, a member of the novel PKC family, may have a distinct isoform-specific role in breast cancer. PKCθ is preferentially expressed in triple-negative breast cancer (TNBC) compared to other breast tumor subtypes. We hypothesized that PRKCQ/PKCθ critically regulates growth and survival of a subset of TNBC cells. Methods: To elucidate the role of PRKCQ/PKCθ in regulating growth and anoikis resistance, we used both gain and loss of function to modulate expression of PRKCQ. We enhanced expression of PKCθ (kinase-active or inactive) in non-transformed breast epithelial cells (MCF-10A) and assessed effects on epidermal growth factor (EGF)- independent growth, anoikis, and migration. We downregulated expression of PKCθ in TNBC cells, and determined effects on in vitro and in vivo growth and survival. TNBC cells were also treated with a small molecule inhibitor to assess requirement for PKCθ kinase activity in the growth of TNBC cells. Results: PRKCQ/PKCθ can promote oncogenic phenotypes when expressed in non-transformed MCF-10A mammary epithelial cells; PRKCQ/PKCθ enhances anchorage-independent survival, growth-factor-independent proliferation, and migration. PKCθ expression promotes retinoblastoma (Rb) phosphorylation and cell-cycle progression under growth factor-deprived conditions that typically induce cell-cycle arrest of MCF-10A breast epithelial cells. -

G Protein-Coupled Receptors in Stem Cell Maintenance and Somatic Reprogramming to Pluripotent Or Cancer Stem Cells
BMB Reports - Manuscript Submission Manuscript Draft Manuscript Number: BMB-14-250 Title: G protein-coupled receptors in stem cell maintenance and somatic reprogramming to pluripotent or cancer stem cells Article Type: Mini Review Keywords: G protein-coupled receptors; stem cell maintenance; somatic reprogramming; cancer stem cells; pluripotent stem cell Corresponding Author: Ssang-Goo Cho Authors: Ssang-Goo Cho1,*, Hye Yeon Choi1, Subbroto Kumar Saha1, Kyeongseok Kim1, Sangsu Kim1, Gwang-Mo Yang1, BongWoo Kim1, Jin-hoi Kim1 Institution: 1Department of Animal Biotechnology, Animal Resources Research Center, and Incurable Disease Animal Model and Stem Cell Institute (IDASI), Konkuk University, 120 Neungdong-ro, Gwangjin-gu, Seoul 143-701, Republic of Korea, UNCORRECTED PROOF 1 G protein-coupled receptors in stem cell maintenance and somatic reprogramming to 2 pluripotent or cancer stem cells 3 4 Hye Yeon Choi, Subbroto Kumar Saha, Kyeongseok Kim, Sangsu Kim, Gwang-Mo 5 Yang, BongWoo Kim, Jin-hoi Kim, and Ssang-Goo Cho 6 7 Department of Animal Biotechnology, Animal Resources Research Center, and 8 Incurable Disease Animal Model and Stem Cell Institute (IDASI), Konkuk University, 9 120 Neungdong-ro, Gwangjin-gu, Seoul 143-701, Republic of Korea 10 * 11 Address correspondence to Ssang-Goo Cho, Department of Animal Biotechnology and 12 Animal Resources Research Center. Konkuk University, 120 Neungdong-ro, Gwangjin- 13 gu, Seoul 143-701, Republic of Korea. Tel: 82-2-450-4207, Fax: 82-2-450-1044, E-mail: 14 [email protected] 15 16 17 18 19 1 UNCORRECTED PROOF 20 Abstract 21 The G protein-coupled receptors (GPCRs) compose the third largest gene family in the 22 human genome, representing more than 800 distinct genes and 3–5% of the human genome. -

Fucosyltransferase Genes on Porcine Chromosome 6Q11 Are Closely Linked to the Blood Group Inhibitor (S) and Escherichia Coli F18 Receptor (ECF18R) Loci
Mammalian Genome 8, 736–741 (1997). © Springer-Verlag New York Inc. 1997 Two a(1,2) fucosyltransferase genes on porcine Chromosome 6q11 are closely linked to the blood group inhibitor (S) and Escherichia coli F18 receptor (ECF18R) loci E. Meijerink,1 R. Fries,1,*P.Vo¨geli,1 J. Masabanda,1 G. Wigger,1 C. Stricker,1 S. Neuenschwander,1 H.U. Bertschinger,2 G. Stranzinger1 1Institute of Animal Science, Swiss Federal Institute of Technology, ETH-Zentrum, CH-8092 Zurich, Switzerland 2Institute of Veterinary Bacteriology, University of Zurich, CH 8057 Zurich, Switzerland Received: 17 February 1997 / Accepted: 30 May 1997 Abstract. The Escherichia coli F18 receptor locus (ECF18R) has fimbriae F107, has been shown to be genetically controlled by the been genetically mapped to the halothane linkage group on porcine host and is inherited as a dominant trait (Bertschinger et al. 1993) Chromosome (Chr) 6. In an attempt to obtain candidate genes for with B being the susceptibility allele and b the resistance allele. this locus, we isolated 5 cosmids containing the a(1,2)fucosyl- The genetic locus for this E. coli F18 receptor (ECF18R) has been transferase genes FUT1, FUT2, and the pseudogene FUT2P from mapped to porcine Chr 6 (SSC6), based on its close linkage to the a porcine genomic library. Mapping by fluorescence in situ hy- S locus and other loci of the halothane (HAL) linkage group (Vo¨- bridization placed all these clones in band q11 of porcine Chr 6 geli et al. 1996). The epistatic S locus suppresses the phenotypic (SSC6q11). Sequence analysis of the cosmids resulted in the char- expression of the A-0 blood group system when being SsSs (Vo¨geli acterization of an open reading frame (ORF), 1098 bp in length, et al. -

NIH Public Access Author Manuscript Cancer J
NIH Public Access Author Manuscript Cancer J. Author manuscript; available in PMC 2013 March 1. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Cancer J. 2012 March ; 18(2): 124±131. doi:10.1097/PPO.0b013e31824b436e. Targeting Mutant BRAF in Melanoma: Current Status and Future Development of Combination Therapy Strategies Ragini Kudchadkar, MD1, Kim H. T. Paraiso, MS2,3, and Keiran S. M. Smalley, PhD1,2,3,* 1Department of Cutaneous Oncology The Moffitt Cancer Center 12902 Magnolia Drive Tampa, FL, 33612, USA 2Department of Molecular Oncology The Moffitt Cancer Center 12902 Magnolia Drive Tampa, FL, 33612, USA 3The Comprehensive Melanoma Research Center The Moffitt Cancer Center 12902 Magnolia Drive Tampa, FL, 33612, USA Abstract The discovery of activating BRAF mutations in ~50% of all melanomas has proved to be a turning point in the therapeutic management of the disseminated disease. In this commentary we review the latest research delineating the role of mutant BRAF in melanoma initiation and progression and discuss the remarkable 10-year journey leading up to the recent FDA-approval of the small molecule BRAF inhibitor vemurafenib. We further outline the most recent findings on the mechanisms that underlie intrinsic and acquired BRAF inhibitor resistance and describe ongoing preclinical and clinical studies designed to delay or abrogate the onset of therapeutic escape. It is hoped that our evolving understanding of melanoma genetics and intracellular signaling coupled with a growing armamentarium of signal transduction inhibitors will lead to significant improvements in the level and durability of therapeutic response in metastatic melanoma. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase