Genome Sequence of the Palaeopolyploid Soybean
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
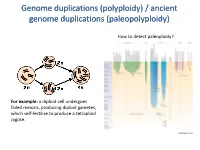
Polyploidy) / Ancient Genome Duplications (Paleopolyploidy
Genome duplications (polyploidy) / ancient genome duplications (paleopolyploidy) How to detect paleoploidy? For example: a diploid cell undergoes failed meiosis, producing diploid gametes, which self-fertilize to produce a tetraploid zygote. Timing of duplication by trees (phylogenetic timing) Phylogenetic timing of duplicates b Paramecium genome duplications Comparison of two scaffolds originating from a common ancestor at the recent WGD Saccharomyces cerevisiae Just before genome duplication Just after genome duplication More time after genome duplication Unaligned view (removing gaps just like in cerev has occurred) Saccharomyces cerevisiae Problem reciprocal gene loss (extreme case); how to solve? Problem reciprocal gene loss (extreme case); how to solve? Just before genome duplication Outgroup! Just after genome duplication Outgroup Just after genome duplication Outgroup More time after genome duplication Outgroup Problem (extreme case); how to solve? Outgroup Outgroup Outgroup Outgroup Outgroup Using other genomes Wong et al. 2002 PNAS Centromeres Vertebrate genome duplication Nature. 2011 Apr 10. [Epub ahead of print] Ancestral polyploidy in seed plants and angiosperms. Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, Depamphilis CW. Flowering plants Flowering MOSS Vertebrates Teleosts S. serevisiae and close relatives Paramecium Reconstructed map of genome duplications allows unprecedented mapping -

A 'David and Goliath'
Current Genomics, 2002, 3, 563-576 563 Nucleolar Dominance: A ‘David and Goliath’ Chromatin Imprinting Process W. Viegas1,*, N. Neves1,2, M. Silva1, A. Caperta1,2, and L. Morais-Cecílio1 1Centro de Botânica Aplicada à Agricultura, Secção de Genética/DBEB, Instituto Superior de Agronomia, 1349-017 Lisboa, 2Departamento de Ciências Biológicas e Naturais, Universidade Lusófona de Humanidades e Tecnologias, Campo Grande 376, 1749-042 Lisboa, Portugal Abstract: Nucleolar dominance is an enigma. The puzzle of differential amphiplasty has remained unresolved since it was first recognised and described in Crepis hybrids by Navashin in 1934. Here we review the body of knowledge that has grown out of the many models that have tried to find the genetic basis for differential rRNA gene expression in hybrids, and present a new interpretation. We propose and discuss a chromatin imprinting model which re-interprets differential amphiplasty in terms of two genomes of differing size occupying a common space within the nucleus, and with heterochromatin as a key player in the scenario. Difference in size between two parental genomes induces an inherited epigenetic mark in the hybrid that allows patterns of chromatin organization to have positional effects on the neighbouring domains. This chromatin imprinting model can be also used to explain complex genomic interactions which transcend nucleolar dominance and which can account for the overall characteristics of hybrids. Gene expression in hybrids, relative to parentage, is seen as being based on the nuclear location of the sequences concerned within their genomic environment, and where the presence of particular repetitive DNA sequences are ‘sensed’, and render silent the adjacent information. -

Gene Loss and Adaptation in Saccharomyces Genomes
Genetics: Published Articles Ahead of Print, published on December 1, 2005 as 10.1534/genetics.105.048900 After the duplication: gene loss and adaptation in Saccharomyces genomes Paul F. Cliften*,1, Robert S. Fulton§, Richard K. Wilson*, §, and Mark Johnston* *Department of Genetics and §Genome Sequencing Center, Washington University School of Medicine, 660 South Euclid Ave., St. Louis, MO, 63110; 1Current address: Department of Biology, Utah State University, 5305 Old Main Hill, Logan, UT, 84322 Running head: Saccharomyces genomic duplications Key words: genomic duplication, comparative sequence analysis, Saccharomyces phylogeny, yeast Corresponding author: Mark Johnston Department of Genetics Campus Box 8232 Washington University Medical School 4566 Scott Ave. St. Louis, MO 63110 TEL: 314-362-2735 FAX: 314-362-2157 [email protected] ABSTRACT The ancient duplication of the Saccharomyces cerevisiae genome and subsequent massive loss of duplicated genes is apparent when it is compared to the genomes of related species that diverged before the duplication event. To learn more about the evolutionary effects of the duplication event, we compared the S. cerevisiae genome to other Saccharomyces genomes. We demonstrate that the whole genome duplication occurred before S. castellii diverged from S. cerevisiae. In addition to more accurately dating the duplication event, this finding allowed us to study the effects of the duplication on two separate lineages. Analyses of the duplication regions of the genomes indicate that most of the duplicated genes (approximately 85%) were lost before the speciation. Only a small amount of paralogous gene loss (4-6%) occurred after speciation. On the other hand, S. castellii appears to have lost several hundred genes that were not retained as duplicated paralogs. -

Iv Gene Duplication and the Evolution of Silenced
Gene Duplication and the Evolution of Silenced Chromatin in Yeasts by Meleah A. Hickman University Program in Genetics and Genomics Duke University Date:_______________________ Approved: ___________________________ Laura Rusche, Advisor ___________________________ Paul Magwene, Chair ___________________________ Fred Dietrich ___________________________ Joe Heitman ___________________________ Beth Sullivan Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the University Program in Genetics and Genomics in the Graduate School of Duke University 2010 i v ABSTRACT Gene Duplication and the Evolution of Silenced Chromatin in Yeasts by Meleah A. Hickman University Program in Genetics and Genomics Duke University Date:_______________________ Approved: ___________________________ Laura Rusche, Advisor ___________________________ Paul Magwene, Chair ___________________________ Fred Dietrich ___________________________ Joe Heitman ___________________________ Beth Sullivan An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the University Program in Genetics and Genomics in the Graduate School of Duke University 2010 Copyright by Meleah A. Hickman 2010 Abstract In Saccharomyces cerevisiae, proper maintenance of haploid cell identity requires the SIR complex to mediate the silenced chromatin found at the cryptic mating-type loci, HML and HMR. This complex consists of Sir2, a histone deacetylase, and the histone binding proteins Sir3 and Sir4. Interestingly, both Sir2 and Sir3 have paralogs from a genome duplication that occurred after the divergence of Saccharomyces and Kluyveromyces species, approximately 100 million years ago. The histone deacetylase HST1 is the paralog of SIR2 and works with the promoter-specific SUM1 complex to repress sporulation and alpha-specific genes. ORC1 is the paralog of SIR3 and is an essential subunit of the Origin Recognition Complex and also recruits SIR proteins to the HM loci. -

Whole-Genome Duplications and Paleopolyploidy Whole-Genome Duplications
Whole-genome duplications and paleopolyploidy Whole-genome duplications AUTOPOLYPLOIDY ALLOPOLYPLOIDY Hegarty and HiscockHegarty 2008, 18 Current Biology Examples of allopolyploid speciation Hegarty and Hiscock 2008, Current Biology 18 Whole-genome duplications of different age time neopolyploidy mesopolyploidy paleopolyploidy time Whole-genome duplications in protozoa •Aury et al. (2006) analyzed the unicellular eukaryote Paramecium tetraurelia • most of 40,000 genes arose through at least 3 successive whole-genome duplications (WGDs) • most recent duplication most likely caused an explosion of speciation events that gave rise to the P. aurelia complex (15 sibling species) • some genes have been lost, some retained • many retained (duplicated) genes do not generate functional innovations but are important because of the gene dosage effect Whole-genome duplications in yeast • genome comparison between two yeast species, Saccharomyces cerevisiae (n=16) and Kluyveromyces waltii (n=8) • each region of K. waltii corresponding to two regions of S. cerevisiae •the S. cerevisiae genome underwent a WGD after the two yeast species diverged • in nearly every case (95%), accelerated evolution was confined to only one of the two paralogues (= one of the paralogues retained an ancestral function, the other was free to evolve more rapidly and acquired a derived function) Kellis et al. 2004, Nature 428 Whole-genome duplications in yeast Kellis et al. 2004, Nature 428 a) after divergence from K. waltii, the Saccharomyces lineage underwent a genome duplication event (2 copies of every gene and chromosome) b) duplicated genes were mutated and some lost c) two copies kept for only a small minority of duplicated genes d) the conserved order of duplicated genes (nos. -

Transposable Elements in Polyploid Evolution Final.Pdf
Transposable elements and polyploid evolution in animals Fernando Rodriguez and Irina R. Arkhipova Address Josephine Bay Paul Center for Comparative Molecular Biology and Evolution, Marine Biological Laboratory, Woods Hole, MA 02543, USA Corresponding author: Arkhipova, Irina R. ([email protected]) Polyploidy in animals is much less common than in plants, where it is thought to be pervasive in all higher plant lineages. Recent studies have highlighted the impact of polyploidization and the associated process of diploidy restoration on the evolution and speciation of selected taxonomic groups in the animal kingdom: from vertebrates represented by salmonid fishes and African clawed frogs to invertebrates represented by parasitic root-knot nematodes and bdelloid rotifers. In this review, we focus on the unique and diverse roles that transposable elements may play in these processes, from marking and diversifying subgenome-specific chromosome sets prior to hybridization, to influencing genome restructuring during rediploidization, to affecting subgenome-specific regulatory evolution, and occasionally providing opportunities for domestication and gene amplification to restore and improve functionality. There is still much to be learned from the future comparative genomic studies of chromosome-sized and haplotype-aware assemblies, and from post-genomic studies elucidating genetic and epigenetic regulatory phenomena across short and long evolutionary distances in the metazoan tree of life. Glossary Polyploidy: having more than the usual diploid set of homologous chromosomes Allopolyploidy: a form of polyploidy that results from interspecific hybridization Autopolyploidy: a form of polyploidy that results from chromosome redoubling in the same species Paleopolyploidy: ancient polyploidy Neopolyploidy: recent polyploidy Homeologs: pairs of genes originated by speciation and brought back together by allopolyploidization Ohnologs: same as homeologs, named after S. -

The Chromosome-Based Lavender Genome Provides New Insights Into
Li et al. Horticulture Research (2021) 8:53 Horticulture Research https://doi.org/10.1038/s41438-021-00490-6 www.nature.com/hortres ARTICLE Open Access The chromosome-based lavender genome provides new insights into Lamiaceae evolution and terpenoid biosynthesis Jingrui Li1,2, Yiming Wang 3,YanmeiDong1,2, Wenying Zhang1,2,DiWang1, Hongtong Bai1,KuiLi3,HuiLi1 and Lei Shi1 Abstract The aromatic shrub Lavandula angustifolia produces various volatile terpenoids that serve as resources for essential oils and function in plant-insect communication. To better understand the genetic basis of the terpenoid diversity in lavender, we present a high-quality reference genome for the Chinese lavender cultivar ‘Jingxun 2’ using PacBio and Hi-C technologies to anchor the 894.50 Mb genome assembly into 27 pseudochromosomes. In addition to the γ triplication event, lavender underwent two rounds of whole-genome duplication (WGD) during the Eocene–Oligocene (29.6 MYA) and Miocene–Pliocene (6.9 MYA) transitions. As a result of tandem duplications and lineage-specific WGDs, gene families related to terpenoid biosynthesis in lavender are substantially expanded compared to those of five other species in Lamiaceae. Many terpenoid biosynthesis transcripts are abundant in glandular trichomes. We further integrated the contents of ecologically functional terpenoids and coexpressed terpenoid biosynthetic genes to construct terpenoid-gene networks. Typical gene clusters, including TPS-TPS, TPS- CYP450, and TPS-BAHD, linked with compounds that primarily function as attractants or repellents, were identified by fl 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; their similar patterns of change during ower development or in response to methyl jasmonate. Comprehensive analysis of the genetic basis of the production of volatiles in lavender could serve as a foundation for future research into lavender evolution, phytochemistry, and ecology. -

Ancestral Grass Karyotype Reconstruction Unravels New Mechanisms of Genome Shuffling As a Source of Plant Evolution
Downloaded from genome.cshlp.org on September 29, 2021 - Published by Cold Spring Harbor Laboratory Press Research Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution Florent Murat,1,4 Jian-Hong Xu,2,4 Eric Tannier,3,4 Michael Abrouk,1 Nicolas Guilhot,1 Caroline Pont,1 Joachim Messing,2,5 and Je´roˆme Salse1,5 1INRA, UMR 1095, Laboratoire Ge´ne´tique, Diversite´ et Ecophysiologie des Ce´re´ales, 63100 Clermont Ferrand, France; 2The Plant Genome Initiative at Rutgers (PGIR), Waksman Institute of Microbiology, Rutgers, The State University of New Jersey, Piscataway, New Jersey 08854, USA; 3INRIA Rhoˆne-Alpes, Universite´ de Lyon 1, CNRS UMR5558, Laboratoire Biome´trie et Biologie E´volutive, 69622 Villeurbanne Cedex, France The comparison of the chromosome numbers of today’s species with common reconstructed paleo-ancestors has led to intense speculation of how chromosomes have been rearranged over time in mammals. However, similar studies in plants with respect to genome evolution as well as molecular mechanisms leading to mosaic synteny blocks have been lacking due to relevant examples of evolutionary zooms from genomic sequences. Such studies require genomes of species that belong to the same family but are diverged to fall into different subfamilies. Our most important crops belong to the family of the grasses, where a number of genomes have now been sequenced. Based on detailed paleogenomics, using inference from n = 5–12 grass ancestral karyotypes (AGKs) in terms of gene content and order, we delineated sequence intervals comprising a complete set of junction break points of orthologous regions from rice, maize, sorghum, and Brachypodium genomes, representing three different subfamilies and different polyploidization events. -

Whole-Genome Duplications and Paleopolyploidy Whole-Genome Duplications in Protozoa
Whole-genome duplications and paleopolyploidy Whole-genome duplications in protozoa •Aury et al. (2006) analyzed the unicellular eukaryote Paramecium tetraurelia • most of 40,000 genes arose through at least 3 successive whole-genome duplications • most recent duplication most likely caused an explosion of speciation events that gave rise to the P. aurelia complex (15 sibling species) • some genes have been lost, some retained • many retained (duplicated) genes do not generate functional innovations but are important because of the gene dosage effect Whole-genome duplications in yeast • genome comparison between two yeast species, Saccharomyces cerevisiae (n=16) and Kluyveromyces waltii (n=8) • each region of K. waltii corresponding to two regions of S. cerevisiae •the S. cerevisiae genome underwent a whole-genome duplication after the two yeast species diverged • in nearly every case (95%), accelerated evolution was confined to only one of the two paralogues (= one of the paralogues retained an ancestral function, the other was free to evolve more rapidly and acquired a derived function) Kellis et al. 2004, Nature 428 Whole-genome duplications in yeast Kellis et al. 2004, Nature 428 a) after divergence from K. waltii, the Saccharomyces lineage underwent a genome duplication event (2 copies of every gene and chromosome) b) duplicated genes underwent were mutated and some lost c) two copies kept for only a small minority of duplicated genes d) the conserved order of duplicated genes (nos. 3-13) across different chromosomal segments e) comparison between genomes of S. cerevisiae and K. waltii reveals the duplicated nature of the S. cerevisiae genome Duplicated nature of the S. -

Multiple Large-Scale Gene and Genome Duplications During the Evolution of Hexapods
Multiple large-scale gene and genome duplications during the evolution of hexapods Zheng Lia,1, George P. Tileyb,c,1, Sally R. Galuskaa, Chris R. Reardona, Thomas I. Kiddera, Rebecca J. Rundella,d, and Michael S. Barkera,2 aDepartment of Ecology and Evolutionary Biology, University of Arizona, Tucson, AZ 85721; bDepartment of Biology, University of Florida, Gainesville, FL 32611; cDepartment of Biology, Duke University, Durham, NC 27708; and dDepartment of Environmental and Forest Biology, State University of New York College of Environmental Science and Forestry, Syracuse, NY 13210 Edited by Michael Freeling, University of California, Berkeley, CA, and approved March 12, 2018 (received for review June 14, 2017) Polyploidy or whole genome duplication (WGD) is a major contrib- than 800,000 described hexapod species (25) are known polyploids utor to genome evolution and diversity. Although polyploidy is (17, 20). However, until recently there were limited data available recognized as an important component of plant evolution, it is to search for evidence of paleopolyploidy among the hexapods generally considered to play a relatively minor role in animal and other animal clades. Thus, the contributions of polyploidy to evolution. Ancient polyploidy is found in the ancestry of some animal evolution and the differences with plant evolution have animals, especially fishes, but there is little evidence for ancient remained unclear. WGDs in other metazoan lineages. Here we use recently published To search for evidence of WGDs among the hexapods, we transcriptomes and genomes from more than 150 species across the leveraged recently released genomic data for the insects (26). insect phylogeny to investigate whether ancient WGDs occurred Combined with additional datasets from public databases, we assembled 128 transcriptomes and 27 genomes with at least one during the evolution of Hexapoda, the most diverse clade of animals. -

A Detailed RFLP Map of Cotton, Gossypium Hirsutum X Gossypium
Copyright 0 1994 by the Genetics Society of America X A Detailed RFLP Map~ of Cotton, Gossypium hirsutum Gossypium barbadense: Chromosome Organization and Evolution in a Disomic Polyploid Genome Alesia J. Reinisch," Jian-mjn Dong,* Curt L. Brubaker,t David M. Stelly," Jonathan F. Wendelt and Andrew H. Paterson" *Department of Soil and Crop Sciences, Texas A&M University, College Station Texas 77843-2474, and +Department of Botany, Iowa State University, Ames, Iowa 50011 Manuscript received January6, 1994 Accepted for publication July12, 1994 ABSTRACT We employ a detailed restriction fragment length polymorphism(RFLP) map to investigate chromo- some organization and evolution in cotton, a disomic polyploid. About 46.2% of nuclear DNA probes detect RFLPs distinguishing Gossypium hirsutum and Gossypium barbadense; and 705 RFLP loci are assembled into 41 linkage groups and 4675 cM. The subgenomic origin (A us. D) of most, and chro- mosomal identityof 14 (of 26), linkage groups is shown. TheA and D subgenomes show similar recom- binational length, suggesting that repetitive DNA in the physically larger A subgenome is recombina- tionally inert. RFLPs are somewhat moreabundant in the D subgenome. Linkage among duplicatedRFLPs reveals 11 pairs of homoeologous chromosomal regions-two appear homosequential,most differ by in- versions, and at least one differs by a translocation. Most homoeologies involve chromosomes from different subgenomes, putatively reflecting the n = 13 to n = 26 polyploidization event of 1.1-1.9 mil- lionyears ago. Several observations suggest that another,earlier, polyploidization event spawned n = 13 cottons, at least 25 million years ago. The cotton genome contains about 400-kb DNA per cM, hence mapbased gene cloning is feasible. -

Paleopolyploidy and Gene Duplication in Soybean and Other Legumes Randy C Shoemaker1, Jessica Schlueter1 and Jeff J Doyle2
Paleopolyploidy and gene duplication in soybean and other legumes Randy C Shoemaker1, Jessica Schlueter1 and Jeff J Doyle2 Two of the most important observations from whole-genome 11 in the phaseoloids [4]. Glycine is an exception to these sequences have been the high rate of gene birth and death and low chromosome numbers, and as such is a rarity among the prevalence of large-scale duplication events, including legumes [4]. The genus is divided into two subgenera, polyploidy. There is also a growing appreciation that polyploidy one of which includes only soybean (Glycine max) and its is more than the sum of the gene duplications it creates, in part wild progenitor (Glycine soja), both of which are annual because polyploidy duplicates the members of entire Asian species with 2n = 40. Members of the subgenus regulatory networks. Thus, it may be important to distinguish Glycine are perennials, with around 25 ‘diploid’ (2n =38or paralogs that are produced by individual gene duplications 40) species confined to Australia and around nine allote- from the homoeologous sequences produced by traploid (2n = 78–80) taxa [5]. Thus, all extant species of (allo)polyploidy. This is not a simple task, for several reasons, Glycine are the products of an ancient genome duplication including the chromosomally cryptic nature of many event, with multiple neopolyploid speciation events hav- duplications and the variable rates of gene evolution. Recent ing been superimposed on this paleopolyploid genome progress has been made in understanding patterns of gene and within the past 50 000 years. A review on legume phy- genome duplication in the legume family, specifically in logenetics is presented by Cronk et al.