The Chromosome-Based Lavender Genome Provides New Insights Into
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
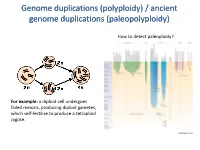
Polyploidy) / Ancient Genome Duplications (Paleopolyploidy
Genome duplications (polyploidy) / ancient genome duplications (paleopolyploidy) How to detect paleoploidy? For example: a diploid cell undergoes failed meiosis, producing diploid gametes, which self-fertilize to produce a tetraploid zygote. Timing of duplication by trees (phylogenetic timing) Phylogenetic timing of duplicates b Paramecium genome duplications Comparison of two scaffolds originating from a common ancestor at the recent WGD Saccharomyces cerevisiae Just before genome duplication Just after genome duplication More time after genome duplication Unaligned view (removing gaps just like in cerev has occurred) Saccharomyces cerevisiae Problem reciprocal gene loss (extreme case); how to solve? Problem reciprocal gene loss (extreme case); how to solve? Just before genome duplication Outgroup! Just after genome duplication Outgroup Just after genome duplication Outgroup More time after genome duplication Outgroup Problem (extreme case); how to solve? Outgroup Outgroup Outgroup Outgroup Outgroup Using other genomes Wong et al. 2002 PNAS Centromeres Vertebrate genome duplication Nature. 2011 Apr 10. [Epub ahead of print] Ancestral polyploidy in seed plants and angiosperms. Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, Depamphilis CW. Flowering plants Flowering MOSS Vertebrates Teleosts S. serevisiae and close relatives Paramecium Reconstructed map of genome duplications allows unprecedented mapping -

A 'David and Goliath'
Current Genomics, 2002, 3, 563-576 563 Nucleolar Dominance: A ‘David and Goliath’ Chromatin Imprinting Process W. Viegas1,*, N. Neves1,2, M. Silva1, A. Caperta1,2, and L. Morais-Cecílio1 1Centro de Botânica Aplicada à Agricultura, Secção de Genética/DBEB, Instituto Superior de Agronomia, 1349-017 Lisboa, 2Departamento de Ciências Biológicas e Naturais, Universidade Lusófona de Humanidades e Tecnologias, Campo Grande 376, 1749-042 Lisboa, Portugal Abstract: Nucleolar dominance is an enigma. The puzzle of differential amphiplasty has remained unresolved since it was first recognised and described in Crepis hybrids by Navashin in 1934. Here we review the body of knowledge that has grown out of the many models that have tried to find the genetic basis for differential rRNA gene expression in hybrids, and present a new interpretation. We propose and discuss a chromatin imprinting model which re-interprets differential amphiplasty in terms of two genomes of differing size occupying a common space within the nucleus, and with heterochromatin as a key player in the scenario. Difference in size between two parental genomes induces an inherited epigenetic mark in the hybrid that allows patterns of chromatin organization to have positional effects on the neighbouring domains. This chromatin imprinting model can be also used to explain complex genomic interactions which transcend nucleolar dominance and which can account for the overall characteristics of hybrids. Gene expression in hybrids, relative to parentage, is seen as being based on the nuclear location of the sequences concerned within their genomic environment, and where the presence of particular repetitive DNA sequences are ‘sensed’, and render silent the adjacent information. -

Gene Loss and Adaptation in Saccharomyces Genomes
Genetics: Published Articles Ahead of Print, published on December 1, 2005 as 10.1534/genetics.105.048900 After the duplication: gene loss and adaptation in Saccharomyces genomes Paul F. Cliften*,1, Robert S. Fulton§, Richard K. Wilson*, §, and Mark Johnston* *Department of Genetics and §Genome Sequencing Center, Washington University School of Medicine, 660 South Euclid Ave., St. Louis, MO, 63110; 1Current address: Department of Biology, Utah State University, 5305 Old Main Hill, Logan, UT, 84322 Running head: Saccharomyces genomic duplications Key words: genomic duplication, comparative sequence analysis, Saccharomyces phylogeny, yeast Corresponding author: Mark Johnston Department of Genetics Campus Box 8232 Washington University Medical School 4566 Scott Ave. St. Louis, MO 63110 TEL: 314-362-2735 FAX: 314-362-2157 [email protected] ABSTRACT The ancient duplication of the Saccharomyces cerevisiae genome and subsequent massive loss of duplicated genes is apparent when it is compared to the genomes of related species that diverged before the duplication event. To learn more about the evolutionary effects of the duplication event, we compared the S. cerevisiae genome to other Saccharomyces genomes. We demonstrate that the whole genome duplication occurred before S. castellii diverged from S. cerevisiae. In addition to more accurately dating the duplication event, this finding allowed us to study the effects of the duplication on two separate lineages. Analyses of the duplication regions of the genomes indicate that most of the duplicated genes (approximately 85%) were lost before the speciation. Only a small amount of paralogous gene loss (4-6%) occurred after speciation. On the other hand, S. castellii appears to have lost several hundred genes that were not retained as duplicated paralogs. -

Iv Gene Duplication and the Evolution of Silenced
Gene Duplication and the Evolution of Silenced Chromatin in Yeasts by Meleah A. Hickman University Program in Genetics and Genomics Duke University Date:_______________________ Approved: ___________________________ Laura Rusche, Advisor ___________________________ Paul Magwene, Chair ___________________________ Fred Dietrich ___________________________ Joe Heitman ___________________________ Beth Sullivan Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the University Program in Genetics and Genomics in the Graduate School of Duke University 2010 i v ABSTRACT Gene Duplication and the Evolution of Silenced Chromatin in Yeasts by Meleah A. Hickman University Program in Genetics and Genomics Duke University Date:_______________________ Approved: ___________________________ Laura Rusche, Advisor ___________________________ Paul Magwene, Chair ___________________________ Fred Dietrich ___________________________ Joe Heitman ___________________________ Beth Sullivan An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the University Program in Genetics and Genomics in the Graduate School of Duke University 2010 Copyright by Meleah A. Hickman 2010 Abstract In Saccharomyces cerevisiae, proper maintenance of haploid cell identity requires the SIR complex to mediate the silenced chromatin found at the cryptic mating-type loci, HML and HMR. This complex consists of Sir2, a histone deacetylase, and the histone binding proteins Sir3 and Sir4. Interestingly, both Sir2 and Sir3 have paralogs from a genome duplication that occurred after the divergence of Saccharomyces and Kluyveromyces species, approximately 100 million years ago. The histone deacetylase HST1 is the paralog of SIR2 and works with the promoter-specific SUM1 complex to repress sporulation and alpha-specific genes. ORC1 is the paralog of SIR3 and is an essential subunit of the Origin Recognition Complex and also recruits SIR proteins to the HM loci. -

Medicinal Plants, Aromatic Herbs and Fragrance Plants in France
> Medicinal plants, aromatic herbs and fragrance plants in France: a small www.ihc2022.org but thriving sector with a strong traditional base and a dynamic research network Annabelle Bergoënd and Joséphine Piasentin In France, medicinal and aromatic plants paced growth. This production is very diverse small areas. Many producers reserve some (MAP) or herbal, medicinal and aromat- and dynamic, leaning on strong traditional areas as a side crop for their main produc- ic plants (HMAP) industries1 traditionally skills and benefiting from new techniques tion. Many of the plants are not referenced refer to culinary herbs, medicinal plants, and high value processing. With a growing in the European common agricultural pol- and plants cultivated for the fragrance and demand for natural products and unfolding icy (CAP) nomenclature. About 75% of the perfume industries. This agricultural sector opportunities for new markets, HMAP is fac- producers cultivate HMAPs to diversify their encompasses several hundred plant species, ing exciting prospects. production. The largest amount of land is as compared with field crops, such as cereals, occupied by fragrance plants, then medici- which may include single species grains. Characteristics of French HMAP nal plants. Aromatic herbs account for less The organic production for these crops is than 10% of the HMAP cultivated area. In significantly larger than traditional crops of Farm features and production are 2018, the estimated average farm was 4 ha global French agriculture. Wild harvest of geographically diverse for MAP production and 17 ha for perfume HMAP continues and is key for a success- The complexity of the production sector for plants. -

Whole-Genome Duplications and Paleopolyploidy Whole-Genome Duplications
Whole-genome duplications and paleopolyploidy Whole-genome duplications AUTOPOLYPLOIDY ALLOPOLYPLOIDY Hegarty and HiscockHegarty 2008, 18 Current Biology Examples of allopolyploid speciation Hegarty and Hiscock 2008, Current Biology 18 Whole-genome duplications of different age time neopolyploidy mesopolyploidy paleopolyploidy time Whole-genome duplications in protozoa •Aury et al. (2006) analyzed the unicellular eukaryote Paramecium tetraurelia • most of 40,000 genes arose through at least 3 successive whole-genome duplications (WGDs) • most recent duplication most likely caused an explosion of speciation events that gave rise to the P. aurelia complex (15 sibling species) • some genes have been lost, some retained • many retained (duplicated) genes do not generate functional innovations but are important because of the gene dosage effect Whole-genome duplications in yeast • genome comparison between two yeast species, Saccharomyces cerevisiae (n=16) and Kluyveromyces waltii (n=8) • each region of K. waltii corresponding to two regions of S. cerevisiae •the S. cerevisiae genome underwent a WGD after the two yeast species diverged • in nearly every case (95%), accelerated evolution was confined to only one of the two paralogues (= one of the paralogues retained an ancestral function, the other was free to evolve more rapidly and acquired a derived function) Kellis et al. 2004, Nature 428 Whole-genome duplications in yeast Kellis et al. 2004, Nature 428 a) after divergence from K. waltii, the Saccharomyces lineage underwent a genome duplication event (2 copies of every gene and chromosome) b) duplicated genes were mutated and some lost c) two copies kept for only a small minority of duplicated genes d) the conserved order of duplicated genes (nos. -

Transposable Elements in Polyploid Evolution Final.Pdf
Transposable elements and polyploid evolution in animals Fernando Rodriguez and Irina R. Arkhipova Address Josephine Bay Paul Center for Comparative Molecular Biology and Evolution, Marine Biological Laboratory, Woods Hole, MA 02543, USA Corresponding author: Arkhipova, Irina R. ([email protected]) Polyploidy in animals is much less common than in plants, where it is thought to be pervasive in all higher plant lineages. Recent studies have highlighted the impact of polyploidization and the associated process of diploidy restoration on the evolution and speciation of selected taxonomic groups in the animal kingdom: from vertebrates represented by salmonid fishes and African clawed frogs to invertebrates represented by parasitic root-knot nematodes and bdelloid rotifers. In this review, we focus on the unique and diverse roles that transposable elements may play in these processes, from marking and diversifying subgenome-specific chromosome sets prior to hybridization, to influencing genome restructuring during rediploidization, to affecting subgenome-specific regulatory evolution, and occasionally providing opportunities for domestication and gene amplification to restore and improve functionality. There is still much to be learned from the future comparative genomic studies of chromosome-sized and haplotype-aware assemblies, and from post-genomic studies elucidating genetic and epigenetic regulatory phenomena across short and long evolutionary distances in the metazoan tree of life. Glossary Polyploidy: having more than the usual diploid set of homologous chromosomes Allopolyploidy: a form of polyploidy that results from interspecific hybridization Autopolyploidy: a form of polyploidy that results from chromosome redoubling in the same species Paleopolyploidy: ancient polyploidy Neopolyploidy: recent polyploidy Homeologs: pairs of genes originated by speciation and brought back together by allopolyploidization Ohnologs: same as homeologs, named after S. -

Chloroplast Genome Analysis of Angiosperms and Phylogenetic Relationships Among
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.05.078212; this version posted May 5, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Chloroplast genome analysis of Angiosperms and phylogenetic relationships among Lamiaceae members with particular reference to teak (Tectona grandis L.f) P. MAHESWARI, C. KUNHIKANNAN AND R. YASODHA* Institute of Forest Genetics and Tree Breeding, Coimbatore 641 002 INDIA *Author for correspondence R. YASODHA, Institute of Forest Genetics and Tree Breeding, Coimbatore, India Telephone: +91 422 2484114; Fax number : +91 422 248549; e.mail: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.05.05.078212; this version posted May 5, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Availability of comprehensive phylogenetic tree for flowering plants which includes many of the economically important crops and trees is one of the essential requirements of plant biologists for diverse applications. It is the first study on the use of chloroplast genome of 3265 Angiosperm taxa to identify evolutionary relationships among the plant species. Sixty genes from chloroplast genome was concatenated and utilized to generate the phylogenetic tree. Overall the phylogeny was in correspondence with Angiosperm Phylogeny Group (APG) IV classification with very few taxa occupying incongruous position either due to ambiguous taxonomy or incorrect identification. Simple sequence repeats (SSRs) were identified from almost all the taxa indicating the possibility of their use in various genetic analyses. -

Chemotaxonomy, Morphology and Chemo Diversity of Scutellaria (Lamiaceae) Species in Zagros, Iran
Journal of Sciences, Islamic Republic of Iran 30(3): 211 - 226 (2019) http://jsciences.ut.ac.ir University of Tehran, ISSN 1016-1104 Chemotaxonomy, Morphology and Chemo Diversity of Scutellaria (Lamiaceae) Species in Zagros, Iran F. Jafari Dehkordi and N. Kharazian* Department of Botany, Faculty of Sciences, Shahrekord University, Shahrekord, Islamic Republic of Iran Received: 2 June 2018 / Revised: 11 May 2019 / Accepted: 26 May 2019 Abstract This study concerns to evaluate the morphological and flavonoid variations, and chemotaxonomy among seven Scutellaria species. The limits of Scutellaria species were disturbed by different factors including hybridization and polymorphism. For this purpose, 39 Scutellaria accessions were collected from different natural habitats of Zagros region, Iran. A total of 15 quantitative and 20 qualitative morphological characters were studied. Leaf flavonoids were extracted using MeOH solution. The flavonoid classes were investigated using thin layer chromatography, column chromatography, UV-spect and LC-MS/MS (liquid chromatography mass spectrometry). To detect the taxonomic status of Scutellaria species, statistical analyses such as cluster, dissimilarity tree, and ordination methods were applied. The results of this research showed five flavonoid classes in different Scutellaria species including isoflavone, flavone, flavanone, flavonol and chalcone. Based on the cluster analysis of flavonoid and morphological data, the members of Scutellaria section Scutellaria were accurately separated from those of Scutellaria section Lupulinaria. Our study revealed a relationship between Scutellaria patonii and Scutellaria multicaulis. Moreover, the trichomes such as strigose, lanate, tomentose, pannous in leaf and stem, petiole, calyx, the form of leaf apex, and inflorescence length were found as diagnostic characters. Based on our results, the flavonoid and morphological markers display the taxonomic status of inter and intra-specific levels in Scutellaria. -

Topical Emulsion Containing Lavandula Stoechas Essential Oil As a Therapeutic Agent for Cutaneous Wound Healing
Article Topical Emulsion Containing Lavandula stoechas Essential Oil as a Therapeutic Agent for Cutaneous Wound Healing Mohamed Nadjib Boukhatem 1,2,* , Henni Chader 3,4, Aicha Houche 1, Faiza Oudjida 5, Fatma Benkebaili 1 and Yahia Hakim 6 1 Department of Biology, Faculty of Life and Natural Sciences, University Blida 1, Blida 09000, Algeria; [email protected] (A.H.); [email protected] (F.B.) 2 Laboratoire Ethnobotanique et Substances Naturelles, Ecole Normale Supérieure, Kouba, Alger 16047, Algeria 3 Laboratoire Pharmaco-Toxicologie, Laboratoire National de Contrôle des Produits Pharmaceutiques (LNCPP), Dély Brahim, Alger 16047, Algeria; [email protected] 4 Faculté de Médecine, Université Ben Youcef Ben Khedda, Alger I, Alger 16000, Algeria 5 Laboratoire Anatomie Pathologique, Centre Hospitalo-Universitaire de Beni Messous, Alger 16206, Algeria; [email protected] 6 Extral-Bio Company, Chiffa, Blida 09000, Algeria; [email protected] * Correspondence: [email protected]; Tel.: +213-664983174 Abstract: Background and objectives: The present research was designed to evaluate the chemical composition of Lavandula stoechas essential oil (EOLS) as well as the in vivo wound-healing property. The chemical composition of EOLS was identified by gas chromatography mass spectrometry. Nine- teen compounds of EOLS were reported. Linalool was identified as the major chemical compound Citation: Boukhatem, M.N.; (24.87%), followed by linalyl acetate (19.10%). EOLS showed a high content of oxygenated com- Chader, H.; Houche, A.; Oudjida, F.; pounds (63.54%). In vivo wound healing activity of the topical cream prepared from EOLS (0.5% w/w) Benkebaili, F.; Hakim, Y. Topical was assessed using a circular excision wound model. -

Lavender Varieties at Chatfield Farms
LAVENDER VARIETIES AT CHATFIELD FARMS Betty’s Blue (L. angustifolia) Flowers: Dark blue compact flowers that bloom once a year Foliage: Gray-green, short, uniform stem length Mature Size: 30 inches with 6-8 inch stems Uses: Best used for crafts due to color and bud retention after harvest. Buena Vista (L. angustifolia) Flowers: Bi-colored, dark and light blue-purple; blooms in late spring and again in the fall Foliage: Green Mature Size: 24-30 inches with 10-12 inch stems Uses: Buena Vista’s sweet fragrance makes it good for culinary use. Also great for landscapes since it blooms twice a year. Dilly-Dilly (L. x intermedia) Flowers: Blue-purple Foliage: Grey-green Mature Size: 12-24 inches Uses: Good for making potpourri, sachets and wands due to strong, pleasant fragrance. High oil content makes this variety good for essential oil extraction as well. Edelweiss (L. x intermedia) Flowers: White flowers with faint blue tint along the calyx bloom once in early summer Foliage: Gray-green Mature Size: 24-30 inches with 18-24 inch stems Uses: Great for the landscape in conjunction with other lavenders due to its white blooms. Also adds variety to crafts such as wreaths and swags. Folgate (L. angustifolia) Flowers: Light blue flowers blooms once in early spring Foliage: Gray-green Mature Size: 30 inches with 8-10 inch stems Uses: Great for culinary use. As one of the earlier bloomers, this variety will let you get a jump on trying out new lavender recipes. Grosso (L. x intermedia) Flowers: Purple flowers bloom once in early summer Foliage: Gray-green Mature Size: 32-36 inches with 20-24 inch stems Uses: Most common variety used in lavender oil production; accounts for approximately 70% of world-wide lavender essential oil production. -

Ethnomedicinal Uses of Genus Lavandula (Lamiaceae) in Turkish
International Journal of Academic and Applied Research (IJAAR) ISSN: 2643-9603 Vol. 4 Issue 2, February – 2020, Pages: 5-16 Ethnomedicinal Uses of Genus Lavandula (Lamiaceae) in Turkish Traditional Medicine Mustafa Eray Bozyel*, Elif Merdamert-Bozyel Department of Biology, Faculty of Arts and Science, Canakkale Onsekiz Mart University, Canakkale, TURKEY *Corresponding author: [email protected] Abstract: Lavandula L. (Lamiaceae) is a widely distributed medicinal plant throughout the world and has been used since ancient time. Popular indications of the several species of this genus include treatment of stomach diseases, headache, inflammation, pains, insomnia, diuretic and expectorant. Phytochemical investigations of Lavandula species have revealed that many components from this genus are highly bioactive. There are many reports on the mentioned ethnomedicinal effects. As a result of the study, the authors found that six Lavandula taxa and two multi-herbal formulas are used as herbal medicine in Turkish traditional medicine. Keywords: Lavandula; Lamiaceae; Medicinal plant; Ethnomedicinal uses; Turkish Traditional Medicine Turkey is located at the intersection of three different phytogeographical regions. These phytogeographical regions 1. INTRODUCTION are European-Siberian (in North Anatolia), the Mediterranean (in Western and Southern Anatolia), and Medicinal plant culture is an indispensable accumulation Irano-Turanian (in Central and South-Eastern Anatolia) [88]. of knowledge depending on observations, and trial and errors These phytogeographical regions of Turkey are the main for centuries. It started with garlic, castor oil and myrrh in cause of the evolution of different species of plants. More the Egyptian Ebers papyrus in 1500 BC. It is a traditional than 12,000 plant taxa grow naturally in Turkey.