1 Novel Insights Into Karyotype Evolution and Whole Genome
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Brooklyn, Cloudland, Melsonby (Gaarraay)
BUSH BLITZ SPECIES DISCOVERY PROGRAM Brooklyn, Cloudland, Melsonby (Gaarraay) Nature Refuges Eubenangee Swamp, Hann Tableland, Melsonby (Gaarraay) National Parks Upper Bridge Creek Queensland 29 April–27 May · 26–27 July 2010 Australian Biological Resources Study What is Contents Bush Blitz? Bush Blitz is a four-year, What is Bush Blitz? 2 multi-million dollar Abbreviations 2 partnership between the Summary 3 Australian Government, Introduction 4 BHP Billiton and Earthwatch Reserves Overview 6 Australia to document plants Methods 11 and animals in selected properties across Australia’s Results 14 National Reserve System. Discussion 17 Appendix A: Species Lists 31 Fauna 32 This innovative partnership Vertebrates 32 harnesses the expertise of many Invertebrates 50 of Australia’s top scientists from Flora 62 museums, herbaria, universities, Appendix B: Threatened Species 107 and other institutions and Fauna 108 organisations across the country. Flora 111 Appendix C: Exotic and Pest Species 113 Fauna 114 Flora 115 Glossary 119 Abbreviations ANHAT Australian Natural Heritage Assessment Tool EPBC Act Environment Protection and Biodiversity Conservation Act 1999 (Commonwealth) NCA Nature Conservation Act 1992 (Queensland) NRS National Reserve System 2 Bush Blitz survey report Summary A Bush Blitz survey was conducted in the Cape Exotic vertebrate pests were not a focus York Peninsula, Einasleigh Uplands and Wet of this Bush Blitz, however the Cane Toad Tropics bioregions of Queensland during April, (Rhinella marina) was recorded in both Cloudland May and July 2010. Results include 1,186 species Nature Refuge and Hann Tableland National added to those known across the reserves. Of Park. Only one exotic invertebrate species was these, 36 are putative species new to science, recorded, the Spiked Awlsnail (Allopeas clavulinus) including 24 species of true bug, 9 species of in Cloudland Nature Refuge. -

Repeated Climate-Linked Host Shifts Have Promoted Diversification in a Temperate Clade of Leaf-Mining Flies
Repeated climate-linked host shifts have promoted SPECIAL FEATURE diversification in a temperate clade of leaf-mining flies Isaac S. Winklera,b,1, Charles Mitterb, and Sonja J. Schefferc aDepartment of Entomology, North Carolina State University, Campus Box 7613, Raleigh, NC 27695-7613; bDepartment of Entomology, University of Maryland, 4112 Plant Sciences Building, College Park, MD 20742; and cSystematic Entomology Laboratory, Plant Science Institute, Agricultural Research Service, United States Department of Agriculture, 10300 Baltimore Avenue, Building 003, Room 231, BARC-West, Beltsville, MD 20705 Edited by Anurag A. Agrawal, Cornell University, Ithaca, NY, and accepted by the Editorial Board July 30, 2009 (received for review May 1, 2009) A central but little-tested prediction of ‘‘escape and radiation’’ ever, there is still little evidence on the degree to which changes coevolution is that colonization of novel, chemically defended host in either plant defense or insect ‘‘offense’’ promote diversifica- plant clades accelerates insect herbivore diversification. That the- tion (7). Progress on the insect side has come from several recent ory, in turn, exemplifies one side of a broader debate about the reports plausibly attributing an instance of significantly elevated relative influence on clade dynamics of intrinsic (biotic) vs. extrinsic insect diversity to a co-occurring shift to a new host taxon (5, 10, (physical-environmental) forces. Here, we use a fossil-calibrated 11). Any single instance of elevated diversification, however, molecular chronogram to compare the effects of a major biotic could reflect other influences that happen to be confounded factor (repeated shift to a chemically divergent host plant clade) with the host shift. -
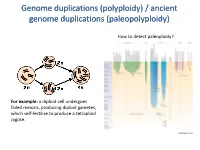
Polyploidy) / Ancient Genome Duplications (Paleopolyploidy
Genome duplications (polyploidy) / ancient genome duplications (paleopolyploidy) How to detect paleoploidy? For example: a diploid cell undergoes failed meiosis, producing diploid gametes, which self-fertilize to produce a tetraploid zygote. Timing of duplication by trees (phylogenetic timing) Phylogenetic timing of duplicates b Paramecium genome duplications Comparison of two scaffolds originating from a common ancestor at the recent WGD Saccharomyces cerevisiae Just before genome duplication Just after genome duplication More time after genome duplication Unaligned view (removing gaps just like in cerev has occurred) Saccharomyces cerevisiae Problem reciprocal gene loss (extreme case); how to solve? Problem reciprocal gene loss (extreme case); how to solve? Just before genome duplication Outgroup! Just after genome duplication Outgroup Just after genome duplication Outgroup More time after genome duplication Outgroup Problem (extreme case); how to solve? Outgroup Outgroup Outgroup Outgroup Outgroup Using other genomes Wong et al. 2002 PNAS Centromeres Vertebrate genome duplication Nature. 2011 Apr 10. [Epub ahead of print] Ancestral polyploidy in seed plants and angiosperms. Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, Depamphilis CW. Flowering plants Flowering MOSS Vertebrates Teleosts S. serevisiae and close relatives Paramecium Reconstructed map of genome duplications allows unprecedented mapping -

1 Noms Vernaculaires : Bengali (Akash Mono,Sonajhuri); English
Fiche présentation arbre : Acacia auriculiformis Plante invasive (ISSG - PIER) : Low risk. A.Cunn. ex Benth. (°) 5 ↑ Utilisations (°) Nom scientifique. Auteur © Benjamin Lisan Noms communs : Acacia auriculiformis Noms vernaculaires : Bengali (akash mono,sonajhuri); English (Japanese acacia,Australian wattle,coast wattle,Darwin black wattle,earleaf acacia,earpod black wattle,earpod wattle,wattle,tan wattle,northern black wattle,Papua wattle); Filipino (auri); Hindi (sonajhuri,kasia,akashmoni,northern black wattle); Indonesian (akasai,kasia,akasia,ki hia); Malay (kasia,akasia kuning); Swahili (mkesia); Thai (krathin-narong); Trade name (Australian wattle); Vietnamese (smach’té:hes) (Source : http://www.worldagroforestry.org/treedb/AFTPDFS/Acacia_auriculiformis.PDF). Noms commerciaux : Synonyme(s) : Distribution, répartition et régions géographiques : Carte de la répartition géographique mondiale. (Source : http://www.cabi.org/isc/datasheet/2157). Latitudes géographiques (°N/ °S): Fourchette d’altitudes : On la trouve entre le niveau de la mer et 1000 m d’altitude (Sourc e : CIRAD). Origine : originaire d'Australie, d'Indonésie et de Papouasie-Nouvelle-Guinée (Source : Wikipedia Fr). Acacia auriculiformis est originaire du Nord de Acacia auriculiformis de 10 ans sur termitière l’Australie, de Papouasie Nouvelle-Guinée et d’Indonésie (Source : CIRAD). (Korhogo) (image CIRAD). Régions d'introduction connues : C’est une espèce qui a été largement plantée de par le monde tropical et subtropical : Inde, Asie du Sud-est, Afrique, Amérique du Sud -

A 'David and Goliath'
Current Genomics, 2002, 3, 563-576 563 Nucleolar Dominance: A ‘David and Goliath’ Chromatin Imprinting Process W. Viegas1,*, N. Neves1,2, M. Silva1, A. Caperta1,2, and L. Morais-Cecílio1 1Centro de Botânica Aplicada à Agricultura, Secção de Genética/DBEB, Instituto Superior de Agronomia, 1349-017 Lisboa, 2Departamento de Ciências Biológicas e Naturais, Universidade Lusófona de Humanidades e Tecnologias, Campo Grande 376, 1749-042 Lisboa, Portugal Abstract: Nucleolar dominance is an enigma. The puzzle of differential amphiplasty has remained unresolved since it was first recognised and described in Crepis hybrids by Navashin in 1934. Here we review the body of knowledge that has grown out of the many models that have tried to find the genetic basis for differential rRNA gene expression in hybrids, and present a new interpretation. We propose and discuss a chromatin imprinting model which re-interprets differential amphiplasty in terms of two genomes of differing size occupying a common space within the nucleus, and with heterochromatin as a key player in the scenario. Difference in size between two parental genomes induces an inherited epigenetic mark in the hybrid that allows patterns of chromatin organization to have positional effects on the neighbouring domains. This chromatin imprinting model can be also used to explain complex genomic interactions which transcend nucleolar dominance and which can account for the overall characteristics of hybrids. Gene expression in hybrids, relative to parentage, is seen as being based on the nuclear location of the sequences concerned within their genomic environment, and where the presence of particular repetitive DNA sequences are ‘sensed’, and render silent the adjacent information. -

Volume 5 Pt 3
Conservation Science W. Aust. 7 (2) : 363–376 (2009) Flora and vegetation of the banded iron formation of the Yilgarn Craton: Robinson Ranges and Mount Gould RACHEL MEISSNER1, GAYNOR OWEN1 & BEN BAYLISS1,2 1 Science Division, Department of Environment and Conservation, PO Box 51, Wanneroo, Western Australia, 6946. Email: [email protected] 2 Avon Natural Diversity Alliance (ANDA), Department of Environment and Conservation, Locked Bag 104,Bentley Delivery Centre WA 6983. ABSTRACT A quadrat based study of the flora and vegetation of the Robinson Ranges and Mount Gould, found 170 taxa including 1 weed taxon. Two priority taxa were recorded and two new taxa identified. Fifty quadrats were established to cover the major geographical, geomorphologic and floristic variation across the hills. Data from 49 of these quadrats were used to define seven community types. Differences in communities were strongly correlated with soil chemistry, elevation, amount of exposed bedrock, surficial rock size and slope. Several communities had restricted distributions. None the plant communities of Robinson Range or Mount Gould are currently in the secure conservation estate. INTRODUCTION by cyclonic activity off the Pilbara coast of Western Australia. Cyclones that cross the coast dissipate and The Robinson Ranges is located in the southern part of develop into rain bearing depressions which may bring the Gascoyne bioregion on the northern edge of the rain into the centre of the state. In addition, thunderstorms Yilgarn Craton. The ranges extend over 200 km, beginning may develop from convectional activity (Curry et al. 1994). near the Great Northern Highway, 140 km north of Winter rainfall is often the result of cold frontal activity Meekatharra, and extending west to Mount Padbury. -

Brisbane Native Plants by Suburb
INDEX - BRISBANE SUBURBS SPECIES LIST Acacia Ridge. ...........15 Chelmer ...................14 Hamilton. .................10 Mayne. .................25 Pullenvale............... 22 Toowong ....................46 Albion .......................25 Chermside West .11 Hawthorne................. 7 McDowall. ..............6 Torwood .....................47 Alderley ....................45 Clayfield ..................14 Heathwood.... 34. Meeandah.............. 2 Queensport ............32 Trinder Park ...............32 Algester.................... 15 Coopers Plains........32 Hemmant. .................32 Merthyr .................7 Annerley ...................32 Coorparoo ................3 Hendra. .................10 Middle Park .........19 Rainworth. ..............47 Underwood. ................41 Anstead ....................17 Corinda. ..................14 Herston ....................5 Milton ...................46 Ransome. ................32 Upper Brookfield .......23 Archerfield ...............32 Highgate Hill. ........43 Mitchelton ...........45 Red Hill.................... 43 Upper Mt gravatt. .......15 Ascot. .......................36 Darra .......................33 Hill End ..................45 Moggill. .................20 Richlands ................34 Ashgrove. ................26 Deagon ....................2 Holland Park........... 3 Moorooka. ............32 River Hills................ 19 Virginia ........................31 Aspley ......................31 Doboy ......................2 Morningside. .........3 Robertson ................42 Auchenflower -

Southern Gulf, Queensland
Biodiversity Summary for NRM Regions Species List What is the summary for and where does it come from? This list has been produced by the Department of Sustainability, Environment, Water, Population and Communities (SEWPC) for the Natural Resource Management Spatial Information System. The list was produced using the AustralianAustralian Natural Natural Heritage Heritage Assessment Assessment Tool Tool (ANHAT), which analyses data from a range of plant and animal surveys and collections from across Australia to automatically generate a report for each NRM region. Data sources (Appendix 2) include national and state herbaria, museums, state governments, CSIRO, Birds Australia and a range of surveys conducted by or for DEWHA. For each family of plant and animal covered by ANHAT (Appendix 1), this document gives the number of species in the country and how many of them are found in the region. It also identifies species listed as Vulnerable, Critically Endangered, Endangered or Conservation Dependent under the EPBC Act. A biodiversity summary for this region is also available. For more information please see: www.environment.gov.au/heritage/anhat/index.html Limitations • ANHAT currently contains information on the distribution of over 30,000 Australian taxa. This includes all mammals, birds, reptiles, frogs and fish, 137 families of vascular plants (over 15,000 species) and a range of invertebrate groups. Groups notnot yet yet covered covered in inANHAT ANHAT are notnot included included in in the the list. list. • The data used come from authoritative sources, but they are not perfect. All species names have been confirmed as valid species names, but it is not possible to confirm all species locations. -

BIODIVERSITY CONSERVATION on the TIWI ISLANDS, NORTHERN TERRITORY: Part 1. Environments and Plants
BIODIVERSITY CONSERVATION ON THE TIWI ISLANDS, NORTHERN TERRITORY: Part 1. Environments and plants Report prepared by John Woinarski, Kym Brennan, Ian Cowie, Raelee Kerrigan and Craig Hempel. Darwin, August 2003 Cover photo: Tall forests dominated by Darwin stringybark Eucalyptus tetrodonta, Darwin woollybutt E. miniata and Melville Island Bloodwood Corymbia nesophila are the principal landscape element across the Tiwi islands (photo: Craig Hempel). i SUMMARY The Tiwi Islands comprise two of Australia’s largest offshore islands - Bathurst (with an area of 1693 km 2) and Melville (5788 km 2) Islands. These are Aboriginal lands lying about 20 km to the north of Darwin, Northern Territory. The islands are of generally low relief with relatively simple geological patterning. They have the highest rainfall in the Northern Territory (to about 2000 mm annual average rainfall in the far north-west of Melville and north of Bathurst). The human population of about 2000 people lives mainly in the three towns of Nguiu, Milakapati and Pirlangimpi. Tall forests dominated by Eucalyptus miniata, E. tetrodonta, and Corymbia nesophila cover about 75% of the island area. These include the best developed eucalypt forests in the Northern Territory. The Tiwi Islands also include nearly 1300 rainforest patches, with floristic composition in many of these patches distinct from that of the Northern Territory mainland. Although the total extent of rainforest on the Tiwi Islands is small (around 160 km 2 ), at an NT level this makes up an unusually high proportion of the landscape and comprises between 6 and 15% of the total NT rainforest extent. The Tiwi Islands also include nearly 200 km 2 of “treeless plains”, a vegetation type largely restricted to these islands. -

Northwest Highlands Bioregion Technical Descriptions
Department of Environment and Science Regional Ecosystem Technical Descriptions Technical descriptions provide a detailed description of the full range in structure and floristic composition of regional ecosystems (e.g. 1.1.10) and their component vegetation communities (e.g. 1.11.10a, 1.11.10b). The descriptions are compiled using site survey data from the Queensland Herbarium’s CORVEG database. Distribution maps, representative images (if available) and the pre-clearing and remnant area (hectares) of each vegetation community derived from the regional ecosystem mapping (spatial) data are included. The technical descriptions should be used in conjunction with the fields from the regional ecosystem description database (REDD) for a full description of the regional ecosystem. Quantitative site data from relatively undisturbed sites are extracted from CORVEG and summarized to provide information specific to each vegetation community. Technical descriptions include the attributes: tree canopy height and cover and native plant species composition of the predominant layer, which are used to assess the remnant status of vegetation under the Vegetation Management Act 1999. However, as technical descriptions reflect the full range in structure and floristic composition across the climatic, natural disturbance and geographic range of the regional ecosystem, local reference sites should be used where possible (Neldner et al. 2005 section 3.3.3). The technical descriptions are subject to review and are updated as additional data becomes available. -

Acacia Thoma Maslin
WATTLE Acacias of Australia Acacia thoma Maslin Source: W orldW ideW attle ver. 2. Source: W orldW ideW attle ver. 2. Published at: w w w .w orldw idew attle.com Published at: w w w .w orldw idew attle.com B.R. Maslin B.R. Maslin Source: W orldW ideW attle ver. 2. Source: W orldW ideW attle ver. 2. Published at: w w w .w orldw idew attle.com Published at: w w w .w orldw idew attle.com B.R. Maslin B.R. Maslin Source: W orldW ideW attle ver. 2. Published at: w w w .w orldw idew attle.com B.R. Maslin Source: W orldW ideW attle ver. 2. Source: W orldW ideW attle ver. 2. Source: W orldW ideW attle ver. 2. Published at: w w w .w orldw idew attle.com Published at: w w w .w orldw idew attle.com Published at: w w w .w orldw idew attle.com B.R. Maslin B.R. Maslin B.R. Maslin Source: W orldW ideW attle ver. 2. Source: W orldW ideW attle ver. 2. Published at: w w w .w orldw idew attle.com Published at: w w w .w orldw idew attle.com B.R. Maslin See illustration. Source: W orldW ideW attle ver. 2. Source: W orldW ideW attle ver. 2. Published at: w w w .w orldw idew attle.com Published at: w w w .w orldw idew attle.com B.R. Maslin B.R. Maslin Acacia thoma occurrence map. O ccurrence map generated via Atlas of Living Australia (https://w w w .ala.org.au). -

Control of Currant Bush (Carissa Ovata) in Developed Brigalow (Acacia Harpophylla) Country
Tropical Grasslands (1998) Volume 32, 259–263 259 Control of currant bush (Carissa ovata) in developed brigalow (Acacia harpophylla) country P.V. BACK can coalesce to cover large areas that signifi- Queensland Beef Industry Institute, Department cantly reduce pasture production. of Primary Industries, Tropical Beef Centre, Ploughing to control brigalow regrowth Rockhampton, Queensland, Australia (Johnson and Back 1974; Scanlan and Anderson 1981) can control currant bush effectively but is very expensive. A more cost-effective treatment is Abstract needed for areas where currant bush dominates in the absence of brigalow regrowth. This paper reports a study designed to test the effectiveness Currant bush (Carissa ovata) is the major native of 6 mechanical methods and 2 herbicide treat- woody weed invading sown buffel grass pastures ments for controlling currant bush in situations in cleared brigalow (Acacia harpophylla) forests where it is the major weed. in Queensland. Stickraking followed by chisel ploughing is a viable alternative to and is more economical than herbicide treatment and blade Materials and methods ploughing for controlling currant bush. Chisel ploughing following stickraking gives good con- Site trol of currant bush with no detrimental effect on existing buffel grass pasture. Stickraking alone is The experiment was carried out on “Tulloch- not sufficient to control currant bush. Ard”, a commercial cattle grazing property 10 km west of Blackwater in central Queensland (23° 33’ S, 148° 44’ E). The original vegetation Introduction comprised a brigalow — blackbutt (Eucalyptus cambageana) scrub with currant bush present in Currant bush (Carissa ovata) is an erect or the understorey, which was cleared and sown to spreading, intricately branched shrub, 1–2 m tall, buffel grass (Cenchrus ciliaris) in 1988.