The Metj Regulon in Gammaproteobacteria Determined by Comparative Genomics Methods Anne M Augustus1 and Leonard D Spicer1,2*
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reporting of Diseases and Conditions Regulation, Amendment, M.R. 289/2014
THE PUBLIC HEALTH ACT LOI SUR LA SANTÉ PUBLIQUE (C.C.S.M. c. P210) (c. P210 de la C.P.L.M.) Reporting of Diseases and Conditions Règlement modifiant le Règlement sur la Regulation, amendment déclaration de maladies et d'affections Regulation 289/2014 Règlement 289/2014 Registered December 23, 2014 Date d'enregistrement : le 23 décembre 2014 Manitoba Regulation 37/2009 amended Modification du R.M. 37/2009 1 The Reporting of Diseases and 1 Le présent règlement modifie le Conditions Regulation , Manitoba Règlement sur la déclaration de maladies et Regulation 37/2009, is amended by this d'affections , R.M. 37/2009. regulation. 2 Schedules A and B are replaced with 2 Les annexes A et B sont remplacées Schedules A and B to this regulation. par les annexes A et B du présent règlement. Coming into force Entrée en vigueur 3 This regulation comes into force on 3 Le présent règlement entre en vigueur January 1, 2015, or on the day it is registered le 1 er janvier 2015 ou à la date de son under The Statutes and Regulations Act , enregistrement en vertu de Loi sur les textes whichever is later. législatifs et réglementaires , si cette date est postérieure. December 19, 2014 Minister of Health/La ministre de la Santé, 19 décembre 2014 Sharon Blady 1 SCHEDULE A (Section 1) 1 The following diseases are diseases requiring contact notification in accordance with the disease-specific protocol. Common name Scientific or technical name of disease or its infectious agent Chancroid Haemophilus ducreyi Chlamydia Chlamydia trachomatis (including Lymphogranuloma venereum (LGV) serovars) Gonorrhea Neisseria gonorrhoeae HIV Human immunodeficiency virus Syphilis Treponema pallidum subspecies pallidum Tuberculosis Mycobacterium tuberculosis Mycobacterium africanum Mycobacterium canetti Mycobacterium caprae Mycobacterium microti Mycobacterium pinnipedii Mycobacterium bovis (excluding M. -

Reportable Disease Surveillance in Virginia, 2013
Reportable Disease Surveillance in Virginia, 2013 Marissa J. Levine, MD, MPH State Health Commissioner Report Production Team: Division of Surveillance and Investigation, Division of Disease Prevention, Division of Environmental Epidemiology, and Division of Immunization Virginia Department of Health Post Office Box 2448 Richmond, Virginia 23218 www.vdh.virginia.gov ACKNOWLEDGEMENT In addition to the employees of the work units listed below, the Office of Epidemiology would like to acknowledge the contributions of all those engaged in disease surveillance and control activities across the state throughout the year. We appreciate the commitment to public health of all epidemiology staff in local and district health departments and the Regional and Central Offices, as well as the conscientious work of nurses, environmental health specialists, infection preventionists, physicians, laboratory staff, and administrators. These persons report or manage disease surveillance data on an ongoing basis and diligently strive to control morbidity in Virginia. This report would not be possible without the efforts of all those who collect and follow up on morbidity reports. Divisions in the Virginia Department of Health Office of Epidemiology Disease Prevention Telephone: 804-864-7964 Environmental Epidemiology Telephone: 804-864-8182 Immunization Telephone: 804-864-8055 Surveillance and Investigation Telephone: 804-864-8141 TABLE OF CONTENTS INTRODUCTION Introduction ......................................................................................................................................1 -
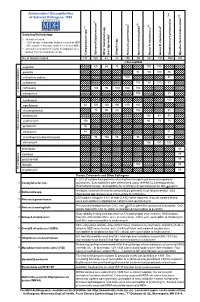
Antimicrobial Susceptibilities of Selected Pathogens, 1999
✔ Antimicrobial Susceptibilities * * † 7 of Selected Pathogens, 1999 8 e † a 2 ✔ ✔ † 3 ✔ † 4 5 6 culosis * r 1 urium spp. m L spp. Sampling Methodology 2 L † all isolates tested * ~ 20% sample of statewide isolates received at MDH spp. ~10% sample of statewide isolates received at MDH Salmonella ** all isolates tested from 7-county metropolitan area oup A streptococci ✔ oup B streptococci r isolates from a normally sterile site r Other (non-typhoidal) G Campylobacter Salmonella typhi Shigella Neisseria gonorrhoeae Neisseria meningitidis G Streptococcus pneumoni Mycobacterium tube No. of Isolates Tested 131 160 43 20 250 55 162 192 559 163 123456 123456% Susceptib123456le 123456123456 123456 123451623456 123451623456 ampicillin 1234561234566012345686 123456 15123456123456 100 100 123456123456 123451623451623451623451623456 123456 penicillin 123456123456123456123456123456 98100 100 76 123456 123456123456123456123456123456123456123456123456 123456 123451623451623451623456 123451623451623456 123456 cefuroxime sodium 123456123456123456123456 100123456123456123456 81 123456 123456123456123456123456123456123456123456123456 123456 cefotaxime 123451623451623451623451623456 100 100100 83 123456 123456123456123456123456123456 123456123456123456123456 123456 123456123456123456123456 ceftriaxone 123456 100 95 100 100 100 123451623451623451623456 -lactam antibiotics 123456123456123456123456123456 123456123456123456123456 β 123451623451623451623451623456 123451623456 123456 meropenem 123456123456123456123456123456 100 123456123456 83 123456 123456123456123456123456123456123456123456123456 -

Microbial NAD Metabolism: Lessons from Comparative Genomics
Dartmouth College Dartmouth Digital Commons Dartmouth Scholarship Faculty Work 9-2009 Microbial NAD Metabolism: Lessons from Comparative Genomics Francesca Gazzaniga Rebecca Stebbins Sheila Z. Chang Mark A. McPeek Dartmouth College Charles Brenner Carver College of Medicine Follow this and additional works at: https://digitalcommons.dartmouth.edu/facoa Part of the Biochemistry Commons, Genetics and Genomics Commons, Medicine and Health Sciences Commons, and the Microbiology Commons Dartmouth Digital Commons Citation Gazzaniga, Francesca; Stebbins, Rebecca; Chang, Sheila Z.; McPeek, Mark A.; and Brenner, Charles, "Microbial NAD Metabolism: Lessons from Comparative Genomics" (2009). Dartmouth Scholarship. 1191. https://digitalcommons.dartmouth.edu/facoa/1191 This Article is brought to you for free and open access by the Faculty Work at Dartmouth Digital Commons. It has been accepted for inclusion in Dartmouth Scholarship by an authorized administrator of Dartmouth Digital Commons. For more information, please contact [email protected]. MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Sept. 2009, p. 529–541 Vol. 73, No. 3 1092-2172/09/$08.00ϩ0 doi:10.1128/MMBR.00042-08 Copyright © 2009, American Society for Microbiology. All Rights Reserved. Microbial NAD Metabolism: Lessons from Comparative Genomics Francesca Gazzaniga,1,2 Rebecca Stebbins,1,2 Sheila Z. Chang,1,2 Mark A. McPeek,2 and Charles Brenner1,3* Departments of Genetics and Biochemistry and Norris Cotton Cancer Center, Dartmouth Medical School, Lebanon, New Hampshire -

The Old Testament Is Dying a Diagnosis and Recommended Treatment 1St Edition Download Free
THE OLD TESTAMENT IS DYING A DIAGNOSIS AND RECOMMENDED TREATMENT 1ST EDITION DOWNLOAD FREE Brent A Strawn | 9780801048883 | | | | | David T. Lamb Strawn offers a few other concrete suggestions about how to save the Old Testament, illustrating several of these by looking at the book of Deuteronomy as a model for second language acquisition. Retrieved 27 August The United States' Centers for Disease Control and Prevention CDC currently recommend that individuals who have been diagnosed and treated for gonorrhea avoid sexual contact with others until at least one week past the final day of treatment in order to prevent the spread of the bacterium. Brent Strawn reminds us of the Old Testament's important role in Christian faith and practice, criticizes current misunderstandings that contribute to its neglect, and offers ways to revitalize its use in the church. None, burning with urinationvaginal dischargedischarge from the penispelvic paintesticular pain [1]. Stunted language learners either: leave faith behind altogether; remain Christian, but look to other resources for how to live their lives; or balkanize in communities that prefer to speak something akin to baby talk — a pidgin-like form of the Old Testament and Bible as a whole — or, worse still, some sort of creole. Geoff, thanks for the reference. Log in. The guest easily identified the passage The Old Testament Is Dying A Diagnosis and Recommended Treatment 1st edition the New Testament, but the Old Testament passage was a swing, and a miss. Instead, our system considers things like how recent a review is and if the reviewer bought the item on Amazon. -

1 Supplementary Material a Major Clade of Prokaryotes with Ancient
Supplementary Material A major clade of prokaryotes with ancient adaptations to life on land Fabia U. Battistuzzi and S. Blair Hedges Data assembly and phylogenetic analyses Protein data set: Amino acid sequences of 25 protein-coding genes (“proteins”) were concatenated in an alignment of 18,586 amino acid sites and 283 species. These proteins included: 15 ribosomal proteins (RPL1, 2, 3, 5, 6, 11, 13, 16; RPS2, 3, 4, 5, 7, 9, 11), four genes (RNA polymerase alpha, beta, and gamma subunits, Transcription antitermination factor NusG) from the functional category of Transcription, three proteins (Elongation factor G, Elongation factor Tu, Translation initiation factor IF2) of the Translation, Ribosomal Structure and Biogenesis functional category, one protein (DNA polymerase III, beta subunit) of the DNA Replication, Recombination and repair category, one protein (Preprotein translocase SecY) of the Cell Motility and Secretion category, and one protein (O-sialoglycoprotein endopeptidase) of the Posttranslational Modification, Protein Turnover, Chaperones category, as annotated in the Cluster of Orthologous Groups (COG) (Tatusov et al. 2001). After removal of multiple strains of the same species, GBlocks 0.91b (Castresana 2000) was applied to each protein in the concatenation to delete poorly aligned sites (i.e., sites with gaps in more than 50% of the species and conserved in less than 50% of the species) with the following parameters: minimum number of sequences for a conserved position: 110, minimum number of sequences for a flank position: 110, maximum number of contiguous non-conserved positions: 32000, allowed gap positions: with half. The signal-to-noise ratio was determined by altering the “minimum length of a block” parameter. -

Nebraska Reportable Disease Chart
Nebraska Reportable Diseases Title 173 Regulations Immediate Notification: Douglas Co. (402)444-7214 (after hrs 402- 444-7000) Lancaster Co (402) 441-8053 (after hrs 402-440-1817) All Other Counties 402-471-1983 Nebraska Public Health Laboratory 24/7 pager 402-888-5588 Labs- automated ELR Labs reporting manually Healthcare providers Updated 5/3/2017 Condition immediate within 7 days monthly immediate within 7 days monthly immediate within 7 days monthly Acinetobacter spp . (all species) x Acquired Immunodeficiency Syndrome (AIDS), as described in 173 NAC 1- 005.01C2 xxx Adenovirus x Aeromonas spp. x Amebae-associated infection (Acanthamoeba spp, Entamoeba histolytica , and Naegleria fowleri )xxx Anthrax (Bacillus anthracis) * ^ xx x Arboviral infections (including, but not limited to, West Nile virus, St. Louis encephalitis virus, Western Equine encephalitis virus, Chikungunya virus, Rift Valley fever virus, Zika and Dengue virus) xxx Astrovirus x Babesiosis (Babesia species) x x x Botulism (Clostridium botulinum )* x x x Brucellosis (Brucella abortus^, B. melitensis^, and B. suis)*^ xx x Burkholderia (Pseudomonas) pseudomallei *^ xx x Campylobacteriosis (Campylobacter species )Do not forward to NPHL for banking or subtyping unless requested xxx Carbapenem-Resistant Enterobacteriaceae (suspected or confirmed)^** xx x Carbon monoxide poisoning (Use break point for non-smokers) xxx Chancroid (Haemophilus ducreyi ) ± x x x Chikungunya virus xxx Citrobacter spp. x Chlamydophila (Chlamydia) pneumoniae x Chlamydia trachomatis infections (nonspecific urethritis, cervicitis, salpingitis, neonatal conjunctivitis, pneumonia)± xxx Cholera (Vibrio cholerae ) ^ x x x Clostridium difficile xxx Coccidiodomycosis (Coccidioides immitis/posadasii )xx x Coronavirus (Not MERS) x Creutzfeldt-Jakob Disease [transmissible spongiform encephalopathy (14-3-3 protein from CSF or any laboratory analysis of brain tissue suggestive of CJD)] xxx Cryptosporidiosis (C. -

University of Ghana College of Basic and Applied Sciences by Shirley Victoria Simpson (10551058) This Thesis Is Submitted To
UNIVERSITY OF GHANA COLLEGE OF BASIC AND APPLIED SCIENCES ISOLATION AND CHARACTERIZATION of Haemophilus ducreyi STRAINS FROM CHILDREN WITH CUTANEOUS LESIONS IN YAWS ENDEMIC REGIONS, GHANA BY SHIRLEY VICTORIA SIMPSON (10551058) THIS THESIS IS SUBMITTED TO THE UNIVERSITY OF GHANA, LEGON IN PARTIAL FULFILLMENT OF THE REQUIREMENT FOR THE AWARD OF MPHIL MOLECULAR CELL BIOLOGY OF INFECTIOUS DISEASES DEGREE JULY, 2017 DECLARATION This is to certify that this thesis is the result of research undertaken by me, Shirley Victoria Simpson towards the award of Master of Philosophy in Molecular Cell Biology of Infectious Diseases in the Department of Biochemistry, Cell and Molecular Biology, School of Biological Sciences, College of Basic And Applied Sciences, University of Ghana. Signature--------------------------------------- Date-------------------------- Shirley Victoria Simpson (Candidate) Signature--------------------------------------- Date----------------------- Prof. Kennedy Kwasi Addo (Supervisor) Signature------------------------------------ Date----------------------- Dr. Lydia Mosi (Co-Supervisor) i ABSTRACT Recent discovery of cutaneous H. ducreyi has complicated the epidemiology of Yaws in endemic countries. Yaws and H. ducreyi ulcers are clinically indistinguishable from each other and some other causes of skin ulcerations. The aim of the study was to isolate and characterize H. ducreyi strains from lesions of children in yaws-endemic areas. Symptomatic patients were first screened with Dual Path Platform (DPP-RDT) Syphilis Screen & Confirm test kit (Chembio, Medford, New York) for yaws. Lesion exudates were tested by culture for H. ducreyi and real-time multiplex PCR assays were used to identify T.p subsp. pertenue DNA and H. ducreyi DNA. Azithromycin (AZT) resistance markers were screened for in T.p subsp. pertenue PCR positives. Bacterial 16S rRNA gene was amplified and sequenced to detect the presence of other pathogenic bacteria. -

Haemophilus Ducreyi Infection and Its Relevance to HIV-1 Acquisition This Information Is Current As of September 25, 2021
Evolution of the Cutaneous Immune Response to Experimental Haemophilus ducreyi Infection and Its Relevance to HIV-1 Acquisition This information is current as of September 25, 2021. Tricia L. Humphreys, Carol T. Schnizlein-Bick, Barry P. Katz, Lee Ann Baldridge, Antoinette F. Hood, Robert A. Hromas and Stanley M. Spinola J Immunol 2002; 169:6316-6323; ; doi: 10.4049/jimmunol.169.11.6316 Downloaded from http://www.jimmunol.org/content/169/11/6316 References This article cites 57 articles, 19 of which you can access for free at: http://www.jimmunol.org/ http://www.jimmunol.org/content/169/11/6316.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 25, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2002 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Evolution of the Cutaneous Immune Response to Experimental Haemophilus ducreyi Infection and Its Relevance to HIV-1 Acquisition1,2 Tricia L. Humphreys,3* Carol T. -

Srfabc Toxin from Xenorhabdus Stockiae Induces Cytotoxicity and Apoptosis in Hela Cells
toxins Article SrfABC Toxin from Xenorhabdus stockiae Induces Cytotoxicity and Apoptosis in HeLa Cells Yawei Sun y, Guoyong Zhang y, Xiaoqing Hou, Shuai Xiao, Xi Yang, Yali Xie, Xiyin Huang, Fan Wang, Xiangtao Mo, Xuezhi Ding, Liqiu Xia and Shengbiao Hu * State Key Laboratory of Developmental Biology of Freshwater Fish, Hunan Provincial Key Laboratory for Microbial Molecular Biology, College of Life Science, Hunan Normal University, Changsha 410081, China; [email protected] (Y.S.); [email protected] (G.Z.); [email protected] (X.H.); [email protected] (S.X.); [email protected] (X.Y.); [email protected] (Y.X.); [email protected] (X.H.); [email protected] (F.W.); [email protected] (X.M.); [email protected] (X.D.); [email protected] (L.X.) * Correspondence: [email protected]; Tel./Fax: 86-0731-88872905 These authors contributed equally to this work. y Received: 21 October 2019; Accepted: 14 November 2019; Published: 22 November 2019 Abstract: Our previous study showed that the srfABC operon, which was originally identified in Salmonella enterica as an SsrB-regulated operon clustered with the flagellar class 2 operon, exhibited significant cytotoxicity against insect midgut CF-203 cells and injectable insecticidal activity against Helicoverpa armigera larvae. The srfABC operon was widely distributed among bacteria, which raises the question of their biological roles in different species. In this study, we investigated the cytotoxic effect of SrfABC toxin on mammalian cell lines. When simultaneously expressed in the Escherichia coli cytoplasm, SrfABC exhibited cytotoxicity against all tested mammalian cancer cell lines (B16, 4T-1, Hep-3B, and HeLa) in a dose-dependent manner. -

Resistance Et Évolution Des Poux Humains Pediculus Humanus
AIX-MARSEILLE UNIVERSITE ECOLE DOCTORALE DES SCIENCES DE LA VIE ET DE LA SANTE FACULTE DE MEDECINE DE MARSEILLE Unité de Recherche Microbes, Evolution, Phylogeny and Infection (MEФI) IHU-Méditerranée infection Thèse présentée pour obtenir le grade de Doctorat d’Aix-Marseille Université Spécialité Pathologie Humaine : Maladies infectieuses Mme Nadia AMANZOUGAGHENE -MEHALLA Resistance et évolution des poux humains Pediculus humanus Soutenue le 05 Juillet 2018 devant le jury : Mme le Professeur Fabienne BREGEON Présidente du jury Mr le Docteur Arezki IZRI Rapporteur Mr le Professeur Lionel ZENNER Rapporteur Mr le Docteur Oleg MEDIANNIKOV Co-directeur de thèse Direction de la Thèse : Mr le Professeur Didier RAOULT Directeur de thèse Mr le Docteur Oleg MEDIANNIKOV Co-directeur de thèse Année Universitaire 2017-2018 Remerciements Je commencerai par remercier le Professeur Didier RAOULT pour m’avoir accueilli au sein de son laboratoire et permis de réaliser ces travaux de recherche sous sa direction et ses conseils avisés. Permettez-moi de vous exprimer mon profond respect. Mes profonds remerciements au Docteur Oleg MEDIANNIKOV pour avoir dirigé ce travail. Vous m’avez donné un support scientifique incontournable et la liberté d’évoluer comme un chercheur. Merci pour m’avoir donné votre temps, votre patience, votre assistance, vos conseils et surtout votre confiance dans toutes les étapes de ce travail. Aussi grande que puisse être ma gratitude, soyez assuré qu'elle ne sera jamais à la hauteur de tous les efforts que vous avez déployé, je vous témoigne le plus profond de mes plaisirs de travailler avec vous. Mes sincères et chaleureux remerciements vont au Professeur Florence FENOLLAR, pour avoir co-dirigé ce travail. -

Diversity Patterns of Bacteriophages Infecting Aggregatibacter and Haemophilus Species Across Clades and Niches
The ISME Journal (2019) 13:2500–2522 https://doi.org/10.1038/s41396-019-0450-8 ARTICLE Diversity patterns of bacteriophages infecting Aggregatibacter and Haemophilus species across clades and niches 1,2,3 4 1,2 1,2 1,2 Szymon P. Szafrański ● Mogens Kilian ● Ines Yang ● Gesa Bei der Wieden ● Andreas Winkel ● 5,6 1,2,3 Jan Hegermann ● Meike Stiesch Received: 28 November 2018 / Revised: 7 March 2019 / Accepted: 26 May 2019 / Published online: 14 June 2019 © The Author(s) 2019. This article is published with open access Abstract Aggregatibacter and Haemophilus species are relevant human commensals and opportunistic pathogens. Consequently, their bacteriophages may have significant impact on human microbial ecology and pathologies. Our aim was to reveal the prevalence and diversity of bacteriophages infecting Aggregatibacter and Haemophilus species that colonize the human body. Genome mining with comparative genomics, screening of clinical isolates, and profiling of metagenomes allowed characterization of 346 phages grouped in 52 clusters and 18 superclusters. Less than 10% of the identified phage clusters were represented by previously characterized phages. Prophage diversity patterns varied significantly for different phage fl 1234567890();,: 1234567890();,: types, host clades, and environmental niches. A more diverse phage community lysogenizes Haemophilus in uenzae and Haemophilus parainfluenzae strains than Aggregatibacter actinomycetemcomitans and “Haemophilus ducreyi”. Co-infections occurred more often in “H. ducreyi”. Phages from Aggregatibacter actinomycetemcomitans preferably lysogenized strains of specific serotype. Prophage patterns shared by subspecies clades of different bacterial species suggest similar ecoevolutionary drivers. Changes in frequencies of DNA uptake signal sequences and guanine–cytosine content reflect phage-host long-term coevolution.