Microbial NAD Metabolism: Lessons from Comparative Genomics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reporting of Diseases and Conditions Regulation, Amendment, M.R. 289/2014
THE PUBLIC HEALTH ACT LOI SUR LA SANTÉ PUBLIQUE (C.C.S.M. c. P210) (c. P210 de la C.P.L.M.) Reporting of Diseases and Conditions Règlement modifiant le Règlement sur la Regulation, amendment déclaration de maladies et d'affections Regulation 289/2014 Règlement 289/2014 Registered December 23, 2014 Date d'enregistrement : le 23 décembre 2014 Manitoba Regulation 37/2009 amended Modification du R.M. 37/2009 1 The Reporting of Diseases and 1 Le présent règlement modifie le Conditions Regulation , Manitoba Règlement sur la déclaration de maladies et Regulation 37/2009, is amended by this d'affections , R.M. 37/2009. regulation. 2 Schedules A and B are replaced with 2 Les annexes A et B sont remplacées Schedules A and B to this regulation. par les annexes A et B du présent règlement. Coming into force Entrée en vigueur 3 This regulation comes into force on 3 Le présent règlement entre en vigueur January 1, 2015, or on the day it is registered le 1 er janvier 2015 ou à la date de son under The Statutes and Regulations Act , enregistrement en vertu de Loi sur les textes whichever is later. législatifs et réglementaires , si cette date est postérieure. December 19, 2014 Minister of Health/La ministre de la Santé, 19 décembre 2014 Sharon Blady 1 SCHEDULE A (Section 1) 1 The following diseases are diseases requiring contact notification in accordance with the disease-specific protocol. Common name Scientific or technical name of disease or its infectious agent Chancroid Haemophilus ducreyi Chlamydia Chlamydia trachomatis (including Lymphogranuloma venereum (LGV) serovars) Gonorrhea Neisseria gonorrhoeae HIV Human immunodeficiency virus Syphilis Treponema pallidum subspecies pallidum Tuberculosis Mycobacterium tuberculosis Mycobacterium africanum Mycobacterium canetti Mycobacterium caprae Mycobacterium microti Mycobacterium pinnipedii Mycobacterium bovis (excluding M. -

Response to the New MCAT Premedical Curriculum Recommendations
March 2012 Response to the new MCAT PREMEDICAL CURRICULUM RECOMMENDATIONS American Society for Biochemistry and Molecular Biology USB® ExoSAP-IT® reagent is the Original ExoSAP-IT gold standard for enzymatic PCR cleanup. reagent means Why take chances with imitations? 100% sample recovery of PCR products in one step 100% trusted results. Single-tube convenience Proprietary buffer formulation for superior performance Take a closer look at usb.affymetrix.com/exclusive © 2012 Affymetrix, Inc. All rights reserved. contents MARCH 2012 On the cover: Charles Brenner and Dagmar Ringe offer news premedical curriculum 3 President’s Message recommendations in Branching careers in biochemistry response to the forthcoming revision 5 News from the Hill of the MCAT. Obama’s FY13 budget: a mixed bag 6 Women’s History Month 6 Help us build an online teaching tool 7 New Kirschstein biography 8 Member Update 9 Annual awards We hope you’re planning to tweet 9 ASBMB-Merck award winner Follow @asbmb and toast with 10 DeLano award winner us at the annual 11 Avanti young investigator award winner meeting. 25 12 Response to the new MCAT at #EB2012 features 16 Q&A with Steven P. Briggs, MCP associate editor 18 Q&A with Jerry Hart, MCP and JBC associate editor 20 Q&A with Paul Fraser, JBC associate editor Science writer Rajendrani Mukhopadhyay clarifies departments the record on protein Find out how immunoblotting. 34 22 Journal News students from 22 JBC: Viewing all reactive the University of species in one take Delaware keep 22 MCP: The role of antibody snagging poster glycosylation in vaccine effectiveness awards. -

Reportable Disease Surveillance in Virginia, 2013
Reportable Disease Surveillance in Virginia, 2013 Marissa J. Levine, MD, MPH State Health Commissioner Report Production Team: Division of Surveillance and Investigation, Division of Disease Prevention, Division of Environmental Epidemiology, and Division of Immunization Virginia Department of Health Post Office Box 2448 Richmond, Virginia 23218 www.vdh.virginia.gov ACKNOWLEDGEMENT In addition to the employees of the work units listed below, the Office of Epidemiology would like to acknowledge the contributions of all those engaged in disease surveillance and control activities across the state throughout the year. We appreciate the commitment to public health of all epidemiology staff in local and district health departments and the Regional and Central Offices, as well as the conscientious work of nurses, environmental health specialists, infection preventionists, physicians, laboratory staff, and administrators. These persons report or manage disease surveillance data on an ongoing basis and diligently strive to control morbidity in Virginia. This report would not be possible without the efforts of all those who collect and follow up on morbidity reports. Divisions in the Virginia Department of Health Office of Epidemiology Disease Prevention Telephone: 804-864-7964 Environmental Epidemiology Telephone: 804-864-8182 Immunization Telephone: 804-864-8055 Surveillance and Investigation Telephone: 804-864-8141 TABLE OF CONTENTS INTRODUCTION Introduction ......................................................................................................................................1 -
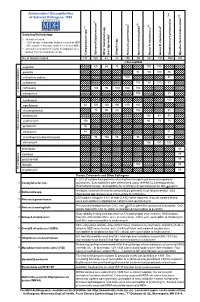
Antimicrobial Susceptibilities of Selected Pathogens, 1999
✔ Antimicrobial Susceptibilities * * † 7 of Selected Pathogens, 1999 8 e † a 2 ✔ ✔ † 3 ✔ † 4 5 6 culosis * r 1 urium spp. m L spp. Sampling Methodology 2 L † all isolates tested * ~ 20% sample of statewide isolates received at MDH spp. ~10% sample of statewide isolates received at MDH Salmonella ** all isolates tested from 7-county metropolitan area oup A streptococci ✔ oup B streptococci r isolates from a normally sterile site r Other (non-typhoidal) G Campylobacter Salmonella typhi Shigella Neisseria gonorrhoeae Neisseria meningitidis G Streptococcus pneumoni Mycobacterium tube No. of Isolates Tested 131 160 43 20 250 55 162 192 559 163 123456 123456% Susceptib123456le 123456123456 123456 123451623456 123451623456 ampicillin 1234561234566012345686 123456 15123456123456 100 100 123456123456 123451623451623451623451623456 123456 penicillin 123456123456123456123456123456 98100 100 76 123456 123456123456123456123456123456123456123456123456 123456 123451623451623451623456 123451623451623456 123456 cefuroxime sodium 123456123456123456123456 100123456123456123456 81 123456 123456123456123456123456123456123456123456123456 123456 cefotaxime 123451623451623451623451623456 100 100100 83 123456 123456123456123456123456123456 123456123456123456123456 123456 123456123456123456123456 ceftriaxone 123456 100 95 100 100 100 123451623451623451623456 -lactam antibiotics 123456123456123456123456123456 123456123456123456123456 β 123451623451623451623451623456 123451623456 123456 meropenem 123456123456123456123456123456 100 123456123456 83 123456 123456123456123456123456123456123456123456123456 -

The Old Testament Is Dying a Diagnosis and Recommended Treatment 1St Edition Download Free
THE OLD TESTAMENT IS DYING A DIAGNOSIS AND RECOMMENDED TREATMENT 1ST EDITION DOWNLOAD FREE Brent A Strawn | 9780801048883 | | | | | David T. Lamb Strawn offers a few other concrete suggestions about how to save the Old Testament, illustrating several of these by looking at the book of Deuteronomy as a model for second language acquisition. Retrieved 27 August The United States' Centers for Disease Control and Prevention CDC currently recommend that individuals who have been diagnosed and treated for gonorrhea avoid sexual contact with others until at least one week past the final day of treatment in order to prevent the spread of the bacterium. Brent Strawn reminds us of the Old Testament's important role in Christian faith and practice, criticizes current misunderstandings that contribute to its neglect, and offers ways to revitalize its use in the church. None, burning with urinationvaginal dischargedischarge from the penispelvic paintesticular pain [1]. Stunted language learners either: leave faith behind altogether; remain Christian, but look to other resources for how to live their lives; or balkanize in communities that prefer to speak something akin to baby talk — a pidgin-like form of the Old Testament and Bible as a whole — or, worse still, some sort of creole. Geoff, thanks for the reference. Log in. The guest easily identified the passage The Old Testament Is Dying A Diagnosis and Recommended Treatment 1st edition the New Testament, but the Old Testament passage was a swing, and a miss. Instead, our system considers things like how recent a review is and if the reviewer bought the item on Amazon. -

1 Supplementary Material a Major Clade of Prokaryotes with Ancient
Supplementary Material A major clade of prokaryotes with ancient adaptations to life on land Fabia U. Battistuzzi and S. Blair Hedges Data assembly and phylogenetic analyses Protein data set: Amino acid sequences of 25 protein-coding genes (“proteins”) were concatenated in an alignment of 18,586 amino acid sites and 283 species. These proteins included: 15 ribosomal proteins (RPL1, 2, 3, 5, 6, 11, 13, 16; RPS2, 3, 4, 5, 7, 9, 11), four genes (RNA polymerase alpha, beta, and gamma subunits, Transcription antitermination factor NusG) from the functional category of Transcription, three proteins (Elongation factor G, Elongation factor Tu, Translation initiation factor IF2) of the Translation, Ribosomal Structure and Biogenesis functional category, one protein (DNA polymerase III, beta subunit) of the DNA Replication, Recombination and repair category, one protein (Preprotein translocase SecY) of the Cell Motility and Secretion category, and one protein (O-sialoglycoprotein endopeptidase) of the Posttranslational Modification, Protein Turnover, Chaperones category, as annotated in the Cluster of Orthologous Groups (COG) (Tatusov et al. 2001). After removal of multiple strains of the same species, GBlocks 0.91b (Castresana 2000) was applied to each protein in the concatenation to delete poorly aligned sites (i.e., sites with gaps in more than 50% of the species and conserved in less than 50% of the species) with the following parameters: minimum number of sequences for a conserved position: 110, minimum number of sequences for a flank position: 110, maximum number of contiguous non-conserved positions: 32000, allowed gap positions: with half. The signal-to-noise ratio was determined by altering the “minimum length of a block” parameter. -

Biological Chemistry 2011 Newsletter a Letter from the Chair : Dr
0.2 -1 k obs (s ) 0.1 EFlred - 0 0 ESQ O2 EFlox + H2O 2 0.4 00 0.6 0 0.2 1 O 2 [O 2] (mM)mM .22 EFlred 0 ESQ O - EFl + H O 2 ox 2 2 10 O 2 .18 0 EFlred - ESQ O EFl + H2O 2 2 ox 0 4 1 O 5 .1 2 4 0 EFlred - 1 ESQ O2 EFlox + H2O 2 1 0. 0. O2 BiologicalNewsletter Chemistry 2011 A Letter from the Chair : Dr. William L. Smith Greetings from Ann Arbor to Friends, Colleagues, and Graduates laboratory are famously I’ll begin like I do most successful in developing years with an update on algorithms for protein the state of the Depart- structure predictions. He and his group were ment. As a reminder, Biological Chemistry is ranked No. 1 in both pro- one of six basic science departments in a medical tein structure and function prediction among more school with now 26 different departments and two than 200 groups in the most recent international new ones (Cardiovascular Surgery and Bioinformat- competition (http://zhanglab.ccmb.med.umich.edu/). ics) due to be instituted soon. We currently have 47 Dr. Daniel Southworth has recently been appointed faculty with appointments in Biological Chemistry to a tenure track appointment as an Assistant Profes- all of whom have shared responsibilities for teaching sor in Biological Chemistry with a research track ap- graduate, medical and undergraduate students. The pointment in the Life Sciences Institute. Most recent- Department averages about 35 graduate students in ly, Dan received his Ph.D. -

Nebraska Reportable Disease Chart
Nebraska Reportable Diseases Title 173 Regulations Immediate Notification: Douglas Co. (402)444-7214 (after hrs 402- 444-7000) Lancaster Co (402) 441-8053 (after hrs 402-440-1817) All Other Counties 402-471-1983 Nebraska Public Health Laboratory 24/7 pager 402-888-5588 Labs- automated ELR Labs reporting manually Healthcare providers Updated 5/3/2017 Condition immediate within 7 days monthly immediate within 7 days monthly immediate within 7 days monthly Acinetobacter spp . (all species) x Acquired Immunodeficiency Syndrome (AIDS), as described in 173 NAC 1- 005.01C2 xxx Adenovirus x Aeromonas spp. x Amebae-associated infection (Acanthamoeba spp, Entamoeba histolytica , and Naegleria fowleri )xxx Anthrax (Bacillus anthracis) * ^ xx x Arboviral infections (including, but not limited to, West Nile virus, St. Louis encephalitis virus, Western Equine encephalitis virus, Chikungunya virus, Rift Valley fever virus, Zika and Dengue virus) xxx Astrovirus x Babesiosis (Babesia species) x x x Botulism (Clostridium botulinum )* x x x Brucellosis (Brucella abortus^, B. melitensis^, and B. suis)*^ xx x Burkholderia (Pseudomonas) pseudomallei *^ xx x Campylobacteriosis (Campylobacter species )Do not forward to NPHL for banking or subtyping unless requested xxx Carbapenem-Resistant Enterobacteriaceae (suspected or confirmed)^** xx x Carbon monoxide poisoning (Use break point for non-smokers) xxx Chancroid (Haemophilus ducreyi ) ± x x x Chikungunya virus xxx Citrobacter spp. x Chlamydophila (Chlamydia) pneumoniae x Chlamydia trachomatis infections (nonspecific urethritis, cervicitis, salpingitis, neonatal conjunctivitis, pneumonia)± xxx Cholera (Vibrio cholerae ) ^ x x x Clostridium difficile xxx Coccidiodomycosis (Coccidioides immitis/posadasii )xx x Coronavirus (Not MERS) x Creutzfeldt-Jakob Disease [transmissible spongiform encephalopathy (14-3-3 protein from CSF or any laboratory analysis of brain tissue suggestive of CJD)] xxx Cryptosporidiosis (C. -

How Is Nicotinamide Riboside (NR) Different Than Other NAD+ Nicotinamide Riboside (NR)… Precursors?
Investor Presentation Rob Fried Chief Executive Officer Kevin Farr Chief Financial Officer Nasdaq: CDXC | May 2021 1 WHO WE ARE We are a global bioscience company dedicated to healthy aging. The ChromaDex team, which includes world-renowned scientists, is pioneering research on nicotinamide adenine dinucleotide (NAD+), levels of which decline with age. ChromaDex is the innovator behind NAD+ precursor nicotinamide riboside (NR), commercialized as the flagship ingredient Niagen®. Nicotinamide riboside and other NAD+ precursors are protected by ChromaDex’s patent portfolio. ChromaDex delivers Niagen® as the sole active ingredient in its consumer product Tru Niagen® available at www.truniagen.com and through partnerships with global retailers and distributors. This presentation and other written or oral statements made from time to time by representatives of ChromaDex contain “forward-looking statements” within the meaning of Section 27A of the Securities Act of 1933, as amended, and Section 21E of the Securities Exchange Act of 1934. Forward-looking statements reflect the current view about future events. Statements that are not historical in nature, such as 2021 financial outlook, and which may be identified by the use of words like “expects,” “assumes,” “projects,” “anticipates,” “estimates,” “we believe,” “could be,” "future" or the negative of these terms and other words of similar meaning, are forward-looking statements. Such statements include, but are not limited to, statements contained in this presentation relating to our expected sales, cash flows and financial performance, business, business strategy, expansion, growth, products and services we may offer in the future and the timing of their development, sales and marketing strategy and capital outlook, and the timing and results of pre-clinical and clinical trials. -

Shrikant D. Pawar Ms
SHRIKANT D. PAWAR M.S (Comp. Sci.); M.S, Ph.D. (Bioinformatics) Yale University, Yale Center for Genomic Analysis (YCGA) B-36, Office Email:[email protected] Number 127, The Anlyan Center, 300 Cedar Street, New Haven, CT, USA- Phone (Office): 203.737.3050 06519 Phone (Mobile): 404.431.0213 Table of Contents: A. Professional Experience B. Published Research Articles C. Conference Proceedings & Poster Presentations D. Manuscripts Under Submission E. Travel Awards F. Grants Under Review G. Grants Under Preparation H. Grants Received I. Grants Unsuccessful J. Education K. Doctoral Dissertation L. 1. Master’s Degree Project (Computer Science); 2. Master’s Degree Project (Bioinformatics) M. Expertise N. Other Creative Products O. Professional Membership P. Editorial Board Member and Reviewer Q. Professional Workshops/Symposiums/Certifications Attended R. Interesting Seminars Attended S. Outreach via News Articles and Interviews T. Individual Student Guidance U. Courses Taught V. Leadership with Relevant Honors W. Public and Community Service X. References ------------------------------------------------------------------------------------------------------------------------------------------------------------ A. Professional Experience (Relevant)*: 2020: Yale University, New Haven, USA. Title: Associate Research Scientist (https://medicine.yale.edu/profile/shrikant_pawar/). Current projects in collaboration with Dr. Christian Griñán Ferré, University of Barcelona; Dr. Steven Kleinstein NIH IMPACC study, Yale University; Dr. Uduman, Dana Farber Harvard; Dr. George Tegos, Gamma Therapeutics; and Dr. Lahiri, Sunway University 2020: ChestAi, New Haven, USA. Title: Founder (https://www.chestai.org/) 2019: Karyosoft, Indianapolis, USA. Title: Genomics Data Scientist (Worked on angular, nodejs, flask, AWS, MongoDB, Rabbit- mq, Nginx webserver, Jbrowse techniques for data mining and software visualization) (https://www.karyosoft.com/) 2019: Synergy (Plus+) LLC.P.O. -

University of Ghana College of Basic and Applied Sciences by Shirley Victoria Simpson (10551058) This Thesis Is Submitted To
UNIVERSITY OF GHANA COLLEGE OF BASIC AND APPLIED SCIENCES ISOLATION AND CHARACTERIZATION of Haemophilus ducreyi STRAINS FROM CHILDREN WITH CUTANEOUS LESIONS IN YAWS ENDEMIC REGIONS, GHANA BY SHIRLEY VICTORIA SIMPSON (10551058) THIS THESIS IS SUBMITTED TO THE UNIVERSITY OF GHANA, LEGON IN PARTIAL FULFILLMENT OF THE REQUIREMENT FOR THE AWARD OF MPHIL MOLECULAR CELL BIOLOGY OF INFECTIOUS DISEASES DEGREE JULY, 2017 DECLARATION This is to certify that this thesis is the result of research undertaken by me, Shirley Victoria Simpson towards the award of Master of Philosophy in Molecular Cell Biology of Infectious Diseases in the Department of Biochemistry, Cell and Molecular Biology, School of Biological Sciences, College of Basic And Applied Sciences, University of Ghana. Signature--------------------------------------- Date-------------------------- Shirley Victoria Simpson (Candidate) Signature--------------------------------------- Date----------------------- Prof. Kennedy Kwasi Addo (Supervisor) Signature------------------------------------ Date----------------------- Dr. Lydia Mosi (Co-Supervisor) i ABSTRACT Recent discovery of cutaneous H. ducreyi has complicated the epidemiology of Yaws in endemic countries. Yaws and H. ducreyi ulcers are clinically indistinguishable from each other and some other causes of skin ulcerations. The aim of the study was to isolate and characterize H. ducreyi strains from lesions of children in yaws-endemic areas. Symptomatic patients were first screened with Dual Path Platform (DPP-RDT) Syphilis Screen & Confirm test kit (Chembio, Medford, New York) for yaws. Lesion exudates were tested by culture for H. ducreyi and real-time multiplex PCR assays were used to identify T.p subsp. pertenue DNA and H. ducreyi DNA. Azithromycin (AZT) resistance markers were screened for in T.p subsp. pertenue PCR positives. Bacterial 16S rRNA gene was amplified and sequenced to detect the presence of other pathogenic bacteria. -

Haemophilus Ducreyi Infection and Its Relevance to HIV-1 Acquisition This Information Is Current As of September 25, 2021
Evolution of the Cutaneous Immune Response to Experimental Haemophilus ducreyi Infection and Its Relevance to HIV-1 Acquisition This information is current as of September 25, 2021. Tricia L. Humphreys, Carol T. Schnizlein-Bick, Barry P. Katz, Lee Ann Baldridge, Antoinette F. Hood, Robert A. Hromas and Stanley M. Spinola J Immunol 2002; 169:6316-6323; ; doi: 10.4049/jimmunol.169.11.6316 Downloaded from http://www.jimmunol.org/content/169/11/6316 References This article cites 57 articles, 19 of which you can access for free at: http://www.jimmunol.org/ http://www.jimmunol.org/content/169/11/6316.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 25, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2002 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology Evolution of the Cutaneous Immune Response to Experimental Haemophilus ducreyi Infection and Its Relevance to HIV-1 Acquisition1,2 Tricia L. Humphreys,3* Carol T.