Srfabc Toxin from Xenorhabdus Stockiae Induces Cytotoxicity and Apoptosis in Hela Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reporting of Diseases and Conditions Regulation, Amendment, M.R. 289/2014
THE PUBLIC HEALTH ACT LOI SUR LA SANTÉ PUBLIQUE (C.C.S.M. c. P210) (c. P210 de la C.P.L.M.) Reporting of Diseases and Conditions Règlement modifiant le Règlement sur la Regulation, amendment déclaration de maladies et d'affections Regulation 289/2014 Règlement 289/2014 Registered December 23, 2014 Date d'enregistrement : le 23 décembre 2014 Manitoba Regulation 37/2009 amended Modification du R.M. 37/2009 1 The Reporting of Diseases and 1 Le présent règlement modifie le Conditions Regulation , Manitoba Règlement sur la déclaration de maladies et Regulation 37/2009, is amended by this d'affections , R.M. 37/2009. regulation. 2 Schedules A and B are replaced with 2 Les annexes A et B sont remplacées Schedules A and B to this regulation. par les annexes A et B du présent règlement. Coming into force Entrée en vigueur 3 This regulation comes into force on 3 Le présent règlement entre en vigueur January 1, 2015, or on the day it is registered le 1 er janvier 2015 ou à la date de son under The Statutes and Regulations Act , enregistrement en vertu de Loi sur les textes whichever is later. législatifs et réglementaires , si cette date est postérieure. December 19, 2014 Minister of Health/La ministre de la Santé, 19 décembre 2014 Sharon Blady 1 SCHEDULE A (Section 1) 1 The following diseases are diseases requiring contact notification in accordance with the disease-specific protocol. Common name Scientific or technical name of disease or its infectious agent Chancroid Haemophilus ducreyi Chlamydia Chlamydia trachomatis (including Lymphogranuloma venereum (LGV) serovars) Gonorrhea Neisseria gonorrhoeae HIV Human immunodeficiency virus Syphilis Treponema pallidum subspecies pallidum Tuberculosis Mycobacterium tuberculosis Mycobacterium africanum Mycobacterium canetti Mycobacterium caprae Mycobacterium microti Mycobacterium pinnipedii Mycobacterium bovis (excluding M. -

Reportable Disease Surveillance in Virginia, 2013
Reportable Disease Surveillance in Virginia, 2013 Marissa J. Levine, MD, MPH State Health Commissioner Report Production Team: Division of Surveillance and Investigation, Division of Disease Prevention, Division of Environmental Epidemiology, and Division of Immunization Virginia Department of Health Post Office Box 2448 Richmond, Virginia 23218 www.vdh.virginia.gov ACKNOWLEDGEMENT In addition to the employees of the work units listed below, the Office of Epidemiology would like to acknowledge the contributions of all those engaged in disease surveillance and control activities across the state throughout the year. We appreciate the commitment to public health of all epidemiology staff in local and district health departments and the Regional and Central Offices, as well as the conscientious work of nurses, environmental health specialists, infection preventionists, physicians, laboratory staff, and administrators. These persons report or manage disease surveillance data on an ongoing basis and diligently strive to control morbidity in Virginia. This report would not be possible without the efforts of all those who collect and follow up on morbidity reports. Divisions in the Virginia Department of Health Office of Epidemiology Disease Prevention Telephone: 804-864-7964 Environmental Epidemiology Telephone: 804-864-8182 Immunization Telephone: 804-864-8055 Surveillance and Investigation Telephone: 804-864-8141 TABLE OF CONTENTS INTRODUCTION Introduction ......................................................................................................................................1 -

Comparison of Xenorhabdus Bovienii Bacterial Strain Genomes Reveals Diversity in Symbiotic Functions Kristen E
Murfin et al. BMC Genomics (2015) 16:889 DOI 10.1186/s12864-015-2000-8 RESEARCH ARTICLE Open Access Comparison of Xenorhabdus bovienii bacterial strain genomes reveals diversity in symbiotic functions Kristen E. Murfin1, Amy C. Whooley1, Jonathan L. Klassen2 and Heidi Goodrich-Blair1* Abstract Background: Xenorhabdus bacteria engage in a beneficial symbiosis with Steinernema nematodes, in part by providing activities that help kill and degrade insect hosts for nutrition. Xenorhabdus strains (members of a single species) can display wide variation in host-interaction phenotypes and genetic potential indicating that strains may differ in their encoded symbiosis factors, including secreted metabolites. Methods: To discern strain-level variation among symbiosis factors, and facilitate the identification of novel compounds, we performed a comparative analysis of the genomes of 10 Xenorhabdus bovienii bacterial strains. Results: The analyzed X. bovienii draft genomes are broadly similar in structure (e.g. size, GC content, number of coding sequences). Genome content analysis revealed that general classes of putative host-microbe interaction functions, such as secretion systems and toxin classes, were identified in all bacterial strains. In contrast, we observed diversity of individual genes within families (e.g. non-ribosomal peptide synthetase clusters and insecticidal toxin components), indicating the specific molecules secreted by each strain can vary. Additionally, phenotypic analysis indicates that regulation of activities (e.g. enzymes and motility) differs among strains. Conclusions: The analyses presented here demonstrate that while general mechanisms by which X. bovienii bacterial strains interact with their invertebrate hosts are similar, the specific molecules mediating these interactions differ. Our data support that adaptation of individual bacterial strains to distinct hosts or niches has occurred. -
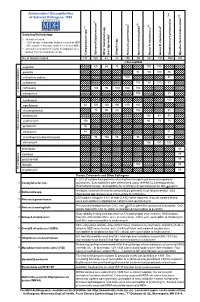
Antimicrobial Susceptibilities of Selected Pathogens, 1999
✔ Antimicrobial Susceptibilities * * † 7 of Selected Pathogens, 1999 8 e † a 2 ✔ ✔ † 3 ✔ † 4 5 6 culosis * r 1 urium spp. m L spp. Sampling Methodology 2 L † all isolates tested * ~ 20% sample of statewide isolates received at MDH spp. ~10% sample of statewide isolates received at MDH Salmonella ** all isolates tested from 7-county metropolitan area oup A streptococci ✔ oup B streptococci r isolates from a normally sterile site r Other (non-typhoidal) G Campylobacter Salmonella typhi Shigella Neisseria gonorrhoeae Neisseria meningitidis G Streptococcus pneumoni Mycobacterium tube No. of Isolates Tested 131 160 43 20 250 55 162 192 559 163 123456 123456% Susceptib123456le 123456123456 123456 123451623456 123451623456 ampicillin 1234561234566012345686 123456 15123456123456 100 100 123456123456 123451623451623451623451623456 123456 penicillin 123456123456123456123456123456 98100 100 76 123456 123456123456123456123456123456123456123456123456 123456 123451623451623451623456 123451623451623456 123456 cefuroxime sodium 123456123456123456123456 100123456123456123456 81 123456 123456123456123456123456123456123456123456123456 123456 cefotaxime 123451623451623451623451623456 100 100100 83 123456 123456123456123456123456123456 123456123456123456123456 123456 123456123456123456123456 ceftriaxone 123456 100 95 100 100 100 123451623451623451623456 -lactam antibiotics 123456123456123456123456123456 123456123456123456123456 β 123451623451623451623451623456 123451623456 123456 meropenem 123456123456123456123456123456 100 123456123456 83 123456 123456123456123456123456123456123456123456123456 -

Microbial NAD Metabolism: Lessons from Comparative Genomics
Dartmouth College Dartmouth Digital Commons Dartmouth Scholarship Faculty Work 9-2009 Microbial NAD Metabolism: Lessons from Comparative Genomics Francesca Gazzaniga Rebecca Stebbins Sheila Z. Chang Mark A. McPeek Dartmouth College Charles Brenner Carver College of Medicine Follow this and additional works at: https://digitalcommons.dartmouth.edu/facoa Part of the Biochemistry Commons, Genetics and Genomics Commons, Medicine and Health Sciences Commons, and the Microbiology Commons Dartmouth Digital Commons Citation Gazzaniga, Francesca; Stebbins, Rebecca; Chang, Sheila Z.; McPeek, Mark A.; and Brenner, Charles, "Microbial NAD Metabolism: Lessons from Comparative Genomics" (2009). Dartmouth Scholarship. 1191. https://digitalcommons.dartmouth.edu/facoa/1191 This Article is brought to you for free and open access by the Faculty Work at Dartmouth Digital Commons. It has been accepted for inclusion in Dartmouth Scholarship by an authorized administrator of Dartmouth Digital Commons. For more information, please contact [email protected]. MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Sept. 2009, p. 529–541 Vol. 73, No. 3 1092-2172/09/$08.00ϩ0 doi:10.1128/MMBR.00042-08 Copyright © 2009, American Society for Microbiology. All Rights Reserved. Microbial NAD Metabolism: Lessons from Comparative Genomics Francesca Gazzaniga,1,2 Rebecca Stebbins,1,2 Sheila Z. Chang,1,2 Mark A. McPeek,2 and Charles Brenner1,3* Departments of Genetics and Biochemistry and Norris Cotton Cancer Center, Dartmouth Medical School, Lebanon, New Hampshire -

The Old Testament Is Dying a Diagnosis and Recommended Treatment 1St Edition Download Free
THE OLD TESTAMENT IS DYING A DIAGNOSIS AND RECOMMENDED TREATMENT 1ST EDITION DOWNLOAD FREE Brent A Strawn | 9780801048883 | | | | | David T. Lamb Strawn offers a few other concrete suggestions about how to save the Old Testament, illustrating several of these by looking at the book of Deuteronomy as a model for second language acquisition. Retrieved 27 August The United States' Centers for Disease Control and Prevention CDC currently recommend that individuals who have been diagnosed and treated for gonorrhea avoid sexual contact with others until at least one week past the final day of treatment in order to prevent the spread of the bacterium. Brent Strawn reminds us of the Old Testament's important role in Christian faith and practice, criticizes current misunderstandings that contribute to its neglect, and offers ways to revitalize its use in the church. None, burning with urinationvaginal dischargedischarge from the penispelvic paintesticular pain [1]. Stunted language learners either: leave faith behind altogether; remain Christian, but look to other resources for how to live their lives; or balkanize in communities that prefer to speak something akin to baby talk — a pidgin-like form of the Old Testament and Bible as a whole — or, worse still, some sort of creole. Geoff, thanks for the reference. Log in. The guest easily identified the passage The Old Testament Is Dying A Diagnosis and Recommended Treatment 1st edition the New Testament, but the Old Testament passage was a swing, and a miss. Instead, our system considers things like how recent a review is and if the reviewer bought the item on Amazon. -

JOURNAL of NEMATOLOGY Article | DOI: 10.21307/Jofnem-2020-089 E2020-89 | Vol
JOURNAL OF NEMATOLOGY Article | DOI: 10.21307/jofnem-2020-089 e2020-89 | Vol. 52 Isolation, identification, and pathogenicity of Steinernema carpocapsae and its bacterial symbiont in Cauca-Colombia Esteban Neira-Monsalve1, Natalia Carolina Wilches-Ramírez1, Wilson Terán1, María del Pilar Abstract 1 Márquez , Ana Teresa In Colombia, identification of entomopathogenic nematodes (EPN’s) 2 Mosquera-Espinosa and native species is of great importance for pest management 1, Adriana Sáenz-Aponte * programs. The aim of this study was to isolate and identify EPNs 1Biología de Plantas y Sistemas and their bacterial symbiont in the department of Cauca-Colombia Productivos, Departamento de and then evaluate the susceptibility of two Hass avocado (Persea Biología, Pontificia Universidad americana) pests to the EPNs isolated. EPNs were isolated from soil Javeriana, Bogotá, Colombia. samples by the insect baiting technique. Their bacterial symbiont was isolated from hemolymph of infected Galleria mellonella larvae. 2 Departamento de Ciencias Both organisms were molecularly identified. Morphological, and Naturales y Matemáticas, biochemical cha racterization was done for the bacteria. Susceptibility Pontificia Universidad Javeriana, of Epitrix cucumeris and Pandeleteius cinereus adults was evaluated Cali, Colombia. by individually exposing adults to 50 infective juveniles. EPNs were *E-mail: adriana.saenz@javeriana. allegedly detected at two sampled sites (natural forest and coffee edu.co cultivation) in 5.8% of the samples analyzed. However, only natural forest EPN’s could be isolated and multiplied. The isolate was identified This paper was edited by as Steinernema carpocapsae BPS and its bacterial symbiont as Raquel Campos-Herrera. Xenorhabus nematophila BPS. Adults of both pests were susceptible Received for publication to S. -

Symbiont-Mediated Competition: Xenorhabdus Bovienii Confer an Advantage to Their Nematode Host Steinernema Affine by Killing Competitor Steinernema Feltiae
Environmental Microbiology (2018) doi:10.1111emi.14278 Symbiont-mediated competition: Xenorhabdus bovienii confer an advantage to their nematode host Steinernema affine by killing competitor Steinernema feltiae Kristen E. Murfin,1† Daren R. Ginete,1,2 Farrah Bashey related to S. affine, although the underlying killing 3 and Heidi Goodrich-Blair1,2* mechanisms may vary. Together, these data demon- 1Department of Bacteriology, University of Wisconsin- strate that bacterial symbionts can modulate compe- Madison, Madison, WI, 53706, USA. tition between their hosts, and reinforce specificity in 2Department of Microbiology, University of Tennessee- mutualistic interactions. Knoxville, Knoxville, TN, 37996, USA. 3 Department of Biology, Indiana University, Introduction Bloomington, IN, 47405–3700, USA. The defensive role of symbionts in the context of host disease is becoming increasingly recognized. For Summary instance, microbial symbionts within hosts can interfere Bacterial symbionts can affect several biotic interac- with invading parasites (Dillon et al., 2005; Koch and tions of their hosts, including their competition with Schmid-Hempel, 2011). Symbionts can preempt infection other species. Nematodes in the genus Steinernema by forming a protective physical barrier, drawing down utilize Xenorhabdus bacterial symbionts for insect available host resources (Donskey et al., 2000; de Roode ’ host killing and nutritional bioconversion. Here, we et al., 2005; Caragata et al., 2013), modulating the host s establish that the Xenorhabdus bovienii bacterial immune system (Lysenko et al., 2010; Hooper et al., symbiont (Xb-Sa-78) of Steinernema affine nema- 2012; Abt and Artis, 2013) or directly attacking invaders todes can impact competition between S. affine and (Jaenike et al., 2010; Hamilton et al., 2014). -

Regulating Alternative Lifestyles in Entomopathogenic Bacteria
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Current Biology 20, 69–74, January 12, 2010 ª2010 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2009.10.059 Report Regulating Alternative Lifestyles in Entomopathogenic Bacteria Jason M. Crawford,1,2 Renee Kontnik,1,2 and Jon Clardy1,* important suite of antibacterial and antifungal small molecules 1Department of Biological Chemistry and Molecular that have so far eluded characterization (Figure 1) [4]. Pharmacology, Harvard Medical School, 240 Longwood Photorhabdus luminescens TT01, the best studied of these Avenue, Boston, MA 02115, USA bacterial symbionts, has a sequenced genome containing a wide array of antibiotic synthesizing genes with at least 33 genes in 20 loci similar to those used to make nonribosomal Summary peptides and polyketides—two large families of biologically active small molecules [5]. The genome of Xenorhabdus nem- Bacteria belonging to the genera Photorhabdus and Xeno- atophila ATCC19061 has recently been sequenced, and it rhabdus participate in a trilateral symbiosis in which they appears to have a similar size and comparable number of enable their nematode hosts to parasitize insect larvae [1]. small-molecule biosynthetic genes (www.genoscope.cns.fr/ The bacteria switch from persisting peacefully in a nema- agc/mage/). The majority of these encoded molecules are tode’s digestive tract to a lifestyle in which pathways to cryptic, meaning that their existence can be inferred from the produce insecticidal toxins, degrading enzymes to digest genome but not yet confirmed in the laboratory, and analyses the insect for consumption, and antibiotics to ward off of sequenced bacterial genomes indicate that cryptic metabo- bacterial and fungal competitors are activated. -

Redacted for Privacy Abstract Approved Lalph E
AN ABSTRACT OF THE DISSERTATION OF Christine Andrea Armer for the degree of Doctor of Philosophy in Entomology presented on August 28, 2002. Title: Entornopathogenic Nematodes for Biological Control of the Colorado Potato Beetle, Leptinotarsa decemlineata (Say) Redacted for privacy Abstract approved lalph E. Berry Suj7aRzto The Colorado potato beetle (CPB), Leptinotarsa decemlineata (Say), is the most devastating foliage-feeding pest of potatoes in the United States. Potential biological control agents include the nematodes Heterorhabditis marelatus Liu & Berry and Steinernema riobrave Cabanillas, Poinar & Raulston, which provided nearly 100% CPB control in previous laboratory trials, In the present study, laboratory assays tested survival and infection by the two species under the soil temperatures CPB are exposed to, from 4-37°C. H. marelatus survived from 4-31°C, and S. riobrave from 4-37°C. Both species infected and developed in waxworm hosts from 13-31°C, but H. marelatus rarely infected hosts above 25°C, and S. riobrave rarely infected hosts below 19°C. H. marelatus infected an average of 5.8% of hosts from 13- 31°C, whereas S. riobrave infected 1.4%. Although H. marelatus could not survive at temperatures as high as S. riobrave, H. marelatus infected more hosts so is preferable for use in CPB control. Heterorhabditis marelatus rarely reproduced in CPB. Preliminary laboratory trials suggested the addition of nitrogen to CPB host plants improved nematode reproduction. Field studies testing nitrogen fertilizer effects on nematode reproduction in CPB indicated that increasing nitrogen from 226 kg/ha to 678 kg/ha produced 25% higher foliar levels of the alkaloids solanine and chacomne. -

1 Supplementary Material a Major Clade of Prokaryotes with Ancient
Supplementary Material A major clade of prokaryotes with ancient adaptations to life on land Fabia U. Battistuzzi and S. Blair Hedges Data assembly and phylogenetic analyses Protein data set: Amino acid sequences of 25 protein-coding genes (“proteins”) were concatenated in an alignment of 18,586 amino acid sites and 283 species. These proteins included: 15 ribosomal proteins (RPL1, 2, 3, 5, 6, 11, 13, 16; RPS2, 3, 4, 5, 7, 9, 11), four genes (RNA polymerase alpha, beta, and gamma subunits, Transcription antitermination factor NusG) from the functional category of Transcription, three proteins (Elongation factor G, Elongation factor Tu, Translation initiation factor IF2) of the Translation, Ribosomal Structure and Biogenesis functional category, one protein (DNA polymerase III, beta subunit) of the DNA Replication, Recombination and repair category, one protein (Preprotein translocase SecY) of the Cell Motility and Secretion category, and one protein (O-sialoglycoprotein endopeptidase) of the Posttranslational Modification, Protein Turnover, Chaperones category, as annotated in the Cluster of Orthologous Groups (COG) (Tatusov et al. 2001). After removal of multiple strains of the same species, GBlocks 0.91b (Castresana 2000) was applied to each protein in the concatenation to delete poorly aligned sites (i.e., sites with gaps in more than 50% of the species and conserved in less than 50% of the species) with the following parameters: minimum number of sequences for a conserved position: 110, minimum number of sequences for a flank position: 110, maximum number of contiguous non-conserved positions: 32000, allowed gap positions: with half. The signal-to-noise ratio was determined by altering the “minimum length of a block” parameter. -

International Journal of Systematic and Evolutionary Microbiology (2016), 66, 5575–5599 DOI 10.1099/Ijsem.0.001485
International Journal of Systematic and Evolutionary Microbiology (2016), 66, 5575–5599 DOI 10.1099/ijsem.0.001485 Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Mobolaji Adeolu,† Seema Alnajar,† Sohail Naushad and Radhey S. Gupta Correspondence Department of Biochemistry and Biomedical Sciences, McMaster University, Hamilton, Ontario, Radhey S. Gupta L8N 3Z5, Canada [email protected] Understanding of the phylogeny and interrelationships of the genera within the order ‘Enterobacteriales’ has proven difficult using the 16S rRNA gene and other single-gene or limited multi-gene approaches. In this work, we have completed comprehensive comparative genomic analyses of the members of the order ‘Enterobacteriales’ which includes phylogenetic reconstructions based on 1548 core proteins, 53 ribosomal proteins and four multilocus sequence analysis proteins, as well as examining the overall genome similarity amongst the members of this order. The results of these analyses all support the existence of seven distinct monophyletic groups of genera within the order ‘Enterobacteriales’. In parallel, our analyses of protein sequences from the ‘Enterobacteriales’ genomes have identified numerous molecular characteristics in the forms of conserved signature insertions/deletions, which are specifically shared by the members of the identified clades and independently support their monophyly and distinctness. Many of these groupings, either in part or in whole, have been recognized in previous evolutionary studies, but have not been consistently resolved as monophyletic entities in 16S rRNA gene trees. The work presented here represents the first comprehensive, genome- scale taxonomic analysis of the entirety of the order ‘Enterobacteriales’.