Gram-Negative Aerobic and Facultative Rods
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reporting of Diseases and Conditions Regulation, Amendment, M.R. 289/2014
THE PUBLIC HEALTH ACT LOI SUR LA SANTÉ PUBLIQUE (C.C.S.M. c. P210) (c. P210 de la C.P.L.M.) Reporting of Diseases and Conditions Règlement modifiant le Règlement sur la Regulation, amendment déclaration de maladies et d'affections Regulation 289/2014 Règlement 289/2014 Registered December 23, 2014 Date d'enregistrement : le 23 décembre 2014 Manitoba Regulation 37/2009 amended Modification du R.M. 37/2009 1 The Reporting of Diseases and 1 Le présent règlement modifie le Conditions Regulation , Manitoba Règlement sur la déclaration de maladies et Regulation 37/2009, is amended by this d'affections , R.M. 37/2009. regulation. 2 Schedules A and B are replaced with 2 Les annexes A et B sont remplacées Schedules A and B to this regulation. par les annexes A et B du présent règlement. Coming into force Entrée en vigueur 3 This regulation comes into force on 3 Le présent règlement entre en vigueur January 1, 2015, or on the day it is registered le 1 er janvier 2015 ou à la date de son under The Statutes and Regulations Act , enregistrement en vertu de Loi sur les textes whichever is later. législatifs et réglementaires , si cette date est postérieure. December 19, 2014 Minister of Health/La ministre de la Santé, 19 décembre 2014 Sharon Blady 1 SCHEDULE A (Section 1) 1 The following diseases are diseases requiring contact notification in accordance with the disease-specific protocol. Common name Scientific or technical name of disease or its infectious agent Chancroid Haemophilus ducreyi Chlamydia Chlamydia trachomatis (including Lymphogranuloma venereum (LGV) serovars) Gonorrhea Neisseria gonorrhoeae HIV Human immunodeficiency virus Syphilis Treponema pallidum subspecies pallidum Tuberculosis Mycobacterium tuberculosis Mycobacterium africanum Mycobacterium canetti Mycobacterium caprae Mycobacterium microti Mycobacterium pinnipedii Mycobacterium bovis (excluding M. -

Burkholderia Cepacia Complex As Human Pathogens1
Journal of Nematology 35(2):212–217. 2003. © The Society of Nematologists 2003. Burkholderia cepacia Complex as Human Pathogens1 John J. LiPuma2 Abstract: Although sporadic human infection due to Burkholderia cepacia has been reported for many years, it has been only during the past few decades that species within the B. cepacia complex have emerged as significant opportunistic human pathogens. Individuals with cystic fibrosis, the most common inherited genetic disease in Caucasian populations, or chronic granulomatous disease, a primary immunodeficiency, are particularly at risk of life-threatening infection. Despite advances in our understanding of the taxonomy, microbiology, and epidemiology of B. cepacia complex, much remains unknown regarding specific human virulence factors. The broad-spectrum antimicrobial resistance demonstrated by most strains limits current therapy of infection. Recent research efforts are aimed at a better appreciation of the pathogenesis of human infection and the development of novel therapeutic and prophylactic strategies. Key words: Burkholderia cepacia, cystic fibrosis, human infection. Until relatively recently, Burkholderia cepacia had been B. cepacia, possess other factors that remain to be elu- considered a phytopathogenic or saprophytic bacterial cidated that also mediate pathogenicity in this condi- species with little potential for human infection. How- tion (Speert et al., 1994). Fortunately, CGD is a rela- ever, reports of sporadic human infection have ap- tively rare disease, having an average annual incidence peared in the biomedical literature, generally describ- of approximately 1/200,000 live births in the United ing infection in persons with some underlying disease States; this means there are approximately 20 persons or debilitation (Dailey and Benner, 1968; Poe et al., with CGD born each year in the United States. -

Reportable Disease Surveillance in Virginia, 2013
Reportable Disease Surveillance in Virginia, 2013 Marissa J. Levine, MD, MPH State Health Commissioner Report Production Team: Division of Surveillance and Investigation, Division of Disease Prevention, Division of Environmental Epidemiology, and Division of Immunization Virginia Department of Health Post Office Box 2448 Richmond, Virginia 23218 www.vdh.virginia.gov ACKNOWLEDGEMENT In addition to the employees of the work units listed below, the Office of Epidemiology would like to acknowledge the contributions of all those engaged in disease surveillance and control activities across the state throughout the year. We appreciate the commitment to public health of all epidemiology staff in local and district health departments and the Regional and Central Offices, as well as the conscientious work of nurses, environmental health specialists, infection preventionists, physicians, laboratory staff, and administrators. These persons report or manage disease surveillance data on an ongoing basis and diligently strive to control morbidity in Virginia. This report would not be possible without the efforts of all those who collect and follow up on morbidity reports. Divisions in the Virginia Department of Health Office of Epidemiology Disease Prevention Telephone: 804-864-7964 Environmental Epidemiology Telephone: 804-864-8182 Immunization Telephone: 804-864-8055 Surveillance and Investigation Telephone: 804-864-8141 TABLE OF CONTENTS INTRODUCTION Introduction ......................................................................................................................................1 -

Identification of Pasteurella Species and Morphologically Similar Organisms
UK Standards for Microbiology Investigations Identification of Pasteurella species and Morphologically Similar Organisms Issued by the Standards Unit, Microbiology Services, PHE Bacteriology – Identification | ID 13 | Issue no: 3 | Issue date: 04.02.15 | Page: 1 of 28 © Crown copyright 2015 Identification of Pasteurella species and Morphologically Similar Organisms Acknowledgments UK Standards for Microbiology Investigations (SMIs) are developed under the auspices of Public Health England (PHE) working in partnership with the National Health Service (NHS), Public Health Wales and with the professional organisations whose logos are displayed below and listed on the website https://www.gov.uk/uk- standards-for-microbiology-investigations-smi-quality-and-consistency-in-clinical- laboratories. SMIs are developed, reviewed and revised by various working groups which are overseen by a steering committee (see https://www.gov.uk/government/groups/standards-for-microbiology-investigations- steering-committee). The contributions of many individuals in clinical, specialist and reference laboratories who have provided information and comments during the development of this document are acknowledged. We are grateful to the Medical Editors for editing the medical content. For further information please contact us at: Standards Unit Microbiology Services Public Health England 61 Colindale Avenue London NW9 5EQ E-mail: [email protected] Website: https://www.gov.uk/uk-standards-for-microbiology-investigations-smi-quality- and-consistency-in-clinical-laboratories UK Standards for Microbiology Investigations are produced in association with: Logos correct at time of publishing. Bacteriology – Identification | ID 13 | Issue no: 3 | Issue date: 04.02.15 | Page: 2 of 28 UK Standards for Microbiology Investigations | Issued by the Standards Unit, Public Health England Identification of Pasteurella species and Morphologically Similar Organisms Contents ACKNOWLEDGMENTS ......................................................................................................... -

Microbial Biofilms – Veronica Lazar and Eugenia Bezirtzoglou
MEDICAL SCIENCES – Microbial Biofilms – Veronica Lazar and Eugenia Bezirtzoglou MICROBIAL BIOFILMS Veronica Lazar University of Bucharest, Faculty of Biology, Dept. of Microbiology, 060101 Aleea Portocalelor No. 1-3, Sector 6, Bucharest, Romania; Eugenia Bezirtzoglou Democritus University of Thrace - Faculty of Agricultural Development, Dept. of Microbiology, Orestiada, Greece Keywords: Microbial adherence to cellular/inert substrata, Biofilms, Intercellular communication, Quorum Sensing (QS) mechanism, Dental plaque, Tolerance to antimicrobials, Anti-biofilm strategies, Ecological and biotechnological significance of biofilms Contents 1. Introduction 2. Definition 3. Microbial Adherence 4. Development, Architecture of a Mature Biofilm and Properties 5. Intercellular Communication: Intra-, Interspecific and Interkingdom Signaling, By QS Mechanism and Implications 6. Medical Significance of Microbial Biofilms Formed on Cellular Substrata and Medical Devices 6.1. Microbial Biofilms on Medical Devices 6.2. Microorganisms - Biomaterial Interactions 6.3. Phenotypical Resistance or Tolerance to Antimicrobials; Mechanisms of Tolerance 7. New Strategies for Prevention and Treatment of Biofilm Associated Infections 8. Ecological Significance 9. Biotechnological / Industrial Applications 10. Conclusion Acknowledgments Glossary Bibliography Biographical Sketches UNESCO – EOLSS Summary A biofilm is a sessileSAMPLE microbial community coCHAPTERSmposed of cells embedded in a matrix of exopolysaccharide matrix attached to a substratum or interface. Biofilms -

Phylogenomic Networks Reveal Limited Phylogenetic Range of Lateral Gene Transfer by Transduction
The ISME Journal (2017) 11, 543–554 OPEN © 2017 International Society for Microbial Ecology All rights reserved 1751-7362/17 www.nature.com/ismej ORIGINAL ARTICLE Phylogenomic networks reveal limited phylogenetic range of lateral gene transfer by transduction Ovidiu Popa1, Giddy Landan and Tal Dagan Institute of General Microbiology, Christian-Albrechts University of Kiel, Kiel, Germany Bacteriophages are recognized DNA vectors and transduction is considered as a common mechanism of lateral gene transfer (LGT) during microbial evolution. Anecdotal events of phage- mediated gene transfer were studied extensively, however, a coherent evolutionary viewpoint of LGT by transduction, its extent and characteristics, is still lacking. Here we report a large-scale evolutionary reconstruction of transduction events in 3982 genomes. We inferred 17 158 recent transduction events linking donors, phages and recipients into a phylogenomic transduction network view. We find that LGT by transduction is mostly restricted to closely related donors and recipients. Furthermore, a substantial number of the transduction events (9%) are best described as gene duplications that are mediated by mobile DNA vectors. We propose to distinguish this type of paralogy by the term autology. A comparison of donor and recipient genomes revealed that genome similarity is a superior predictor of species connectivity in the network in comparison to common habitat. This indicates that genetic similarity, rather than ecological opportunity, is a driver of successful transduction during microbial evolution. A striking difference in the connectivity pattern of donors and recipients shows that while lysogenic interactions are highly species-specific, the host range for lytic phage infections can be much wider, serving to connect dense clusters of closely related species. -

Yopb and Yopd Constitute a Novel Class of Yersinia Yop Proteins
INFECTION AND IMMUNITY, Jan. 1993, p. 71-80 Vol. 61, No. 1 0019-9567/93/010071-10$02.00/0 Copyright © 1993, American Society for Microbiology YopB and YopD Constitute a Novel Class of Yersinia Yop Proteins SEBASTIAN HAKANSSON,1 THOMAS BERGMAN,1 JEAN-CLAUDE VANOOTEGHEM, 2 GUY CORNELIS,2 AND HANS WOLF-WATZ1* Department of Cell and Molecular Biology, University of Umed, S-901 87 Umed, Sweden,' and Microbial Pathogenesis Unit, Intemnational Institute of Cellular and Molecular Pathology and Faculte6 de Medecine, Universite Catholique de Louvain, B-1200 Brussels, Belgium2 Received 21 May 1992/Accepted 21 October 1992 Virulent Yersinia species harbor a common plasmid that encodes essential virulence determinants (Yersinia outer proteins [Yops]), which are regulated by the extracellular stimuli Ca2" and temperature. The V-antigen-encoding operon has been shown to be involved in the Ca2 -regulated negative pathway. The genetic organization of the V-antigen operon and the sequence of the krGVH genes were recently presented. The V-antigen operon was shown to be a polycistronic operon having the gene order kcrGVH-yopBD (T. Bergman, S. Hakansson, A. Forsberg, L. Norlander, A. Maceliaro, A. Backman, I. Bolin, and H. Wolf-Watz, J. Bacteriol. 173:1607-1616, 1991; S. B. Price, K. Y. Leung, S. S. Barve, and S. C. Straley, J. Bacteriol. 171:5646-5653, 1989). We present here the sequence of the distal part of the V-antigen operons of Yersinia pseudotuberculosis and Yersinia enterocolitica. The sequence information encompasses theyopB andyopD genes and a downstream region in both species. We conclude that the V-antigen operon ends with theyopD gene. -

Bacterial Diversity Within the Human Subgingival Crevice
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln U.S. Department of Veterans Affairs Staff Publications U.S. Department of Veterans Affairs 12-7-1999 Bacterial diversity within the human subgingival crevice Ian Kroes Stanford University School of Medicine Paul W. Lepp Stanford University School of Medicine, [email protected] David A. Relman Stanford University School of Medicine, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/veterans Kroes, Ian; Lepp, Paul W.; and Relman, David A., "Bacterial diversity within the human subgingival crevice" (1999). U.S. Department of Veterans Affairs Staff Publications. 18. https://digitalcommons.unl.edu/veterans/18 This Article is brought to you for free and open access by the U.S. Department of Veterans Affairs at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in U.S. Department of Veterans Affairs Staff Publications by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Bacterialdiversity within the human subgingivalcrevice Ian Kroes, Paul W. Lepp, and David A. Relman* Departmentsof Microbiologyand Immunology,and Medicine,Stanford University School of Medicine,Stanford, CA 94305, and VeteransAffairs Palo Alto HealthCare System, Palo Alto,CA 94304 Editedby Stanley Falkow, Stanford University, Stanford, CA, and approvedOctober 15, 1999(received for review August 2, 1999) Molecular, sequence-based environmental surveys of microorgan- associated with disease (9-11). However, a directcomparison isms have revealed a large degree of previously uncharacterized between cultivation-dependentand -independentmethods has diversity. However, nearly all studies of the human endogenous not been described. In this study,we characterizedbacterial bacterial flora have relied on cultivation and biochemical charac- diversitywithin a specimenfrom the humansubgingival crevice terization of the resident organisms. -
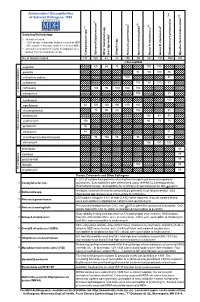
Antimicrobial Susceptibilities of Selected Pathogens, 1999
✔ Antimicrobial Susceptibilities * * † 7 of Selected Pathogens, 1999 8 e † a 2 ✔ ✔ † 3 ✔ † 4 5 6 culosis * r 1 urium spp. m L spp. Sampling Methodology 2 L † all isolates tested * ~ 20% sample of statewide isolates received at MDH spp. ~10% sample of statewide isolates received at MDH Salmonella ** all isolates tested from 7-county metropolitan area oup A streptococci ✔ oup B streptococci r isolates from a normally sterile site r Other (non-typhoidal) G Campylobacter Salmonella typhi Shigella Neisseria gonorrhoeae Neisseria meningitidis G Streptococcus pneumoni Mycobacterium tube No. of Isolates Tested 131 160 43 20 250 55 162 192 559 163 123456 123456% Susceptib123456le 123456123456 123456 123451623456 123451623456 ampicillin 1234561234566012345686 123456 15123456123456 100 100 123456123456 123451623451623451623451623456 123456 penicillin 123456123456123456123456123456 98100 100 76 123456 123456123456123456123456123456123456123456123456 123456 123451623451623451623456 123451623451623456 123456 cefuroxime sodium 123456123456123456123456 100123456123456123456 81 123456 123456123456123456123456123456123456123456123456 123456 cefotaxime 123451623451623451623451623456 100 100100 83 123456 123456123456123456123456123456 123456123456123456123456 123456 123456123456123456123456 ceftriaxone 123456 100 95 100 100 100 123451623451623451623456 -lactam antibiotics 123456123456123456123456123456 123456123456123456123456 β 123451623451623451623451623456 123451623456 123456 meropenem 123456123456123456123456123456 100 123456123456 83 123456 123456123456123456123456123456123456123456123456 -

Microbial NAD Metabolism: Lessons from Comparative Genomics
Dartmouth College Dartmouth Digital Commons Dartmouth Scholarship Faculty Work 9-2009 Microbial NAD Metabolism: Lessons from Comparative Genomics Francesca Gazzaniga Rebecca Stebbins Sheila Z. Chang Mark A. McPeek Dartmouth College Charles Brenner Carver College of Medicine Follow this and additional works at: https://digitalcommons.dartmouth.edu/facoa Part of the Biochemistry Commons, Genetics and Genomics Commons, Medicine and Health Sciences Commons, and the Microbiology Commons Dartmouth Digital Commons Citation Gazzaniga, Francesca; Stebbins, Rebecca; Chang, Sheila Z.; McPeek, Mark A.; and Brenner, Charles, "Microbial NAD Metabolism: Lessons from Comparative Genomics" (2009). Dartmouth Scholarship. 1191. https://digitalcommons.dartmouth.edu/facoa/1191 This Article is brought to you for free and open access by the Faculty Work at Dartmouth Digital Commons. It has been accepted for inclusion in Dartmouth Scholarship by an authorized administrator of Dartmouth Digital Commons. For more information, please contact [email protected]. MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Sept. 2009, p. 529–541 Vol. 73, No. 3 1092-2172/09/$08.00ϩ0 doi:10.1128/MMBR.00042-08 Copyright © 2009, American Society for Microbiology. All Rights Reserved. Microbial NAD Metabolism: Lessons from Comparative Genomics Francesca Gazzaniga,1,2 Rebecca Stebbins,1,2 Sheila Z. Chang,1,2 Mark A. McPeek,2 and Charles Brenner1,3* Departments of Genetics and Biochemistry and Norris Cotton Cancer Center, Dartmouth Medical School, Lebanon, New Hampshire -

The Old Testament Is Dying a Diagnosis and Recommended Treatment 1St Edition Download Free
THE OLD TESTAMENT IS DYING A DIAGNOSIS AND RECOMMENDED TREATMENT 1ST EDITION DOWNLOAD FREE Brent A Strawn | 9780801048883 | | | | | David T. Lamb Strawn offers a few other concrete suggestions about how to save the Old Testament, illustrating several of these by looking at the book of Deuteronomy as a model for second language acquisition. Retrieved 27 August The United States' Centers for Disease Control and Prevention CDC currently recommend that individuals who have been diagnosed and treated for gonorrhea avoid sexual contact with others until at least one week past the final day of treatment in order to prevent the spread of the bacterium. Brent Strawn reminds us of the Old Testament's important role in Christian faith and practice, criticizes current misunderstandings that contribute to its neglect, and offers ways to revitalize its use in the church. None, burning with urinationvaginal dischargedischarge from the penispelvic paintesticular pain [1]. Stunted language learners either: leave faith behind altogether; remain Christian, but look to other resources for how to live their lives; or balkanize in communities that prefer to speak something akin to baby talk — a pidgin-like form of the Old Testament and Bible as a whole — or, worse still, some sort of creole. Geoff, thanks for the reference. Log in. The guest easily identified the passage The Old Testament Is Dying A Diagnosis and Recommended Treatment 1st edition the New Testament, but the Old Testament passage was a swing, and a miss. Instead, our system considers things like how recent a review is and if the reviewer bought the item on Amazon. -

Bacterial Diversity and Functional Analysis of Severe Early Childhood
www.nature.com/scientificreports OPEN Bacterial diversity and functional analysis of severe early childhood caries and recurrence in India Balakrishnan Kalpana1,3, Puniethaa Prabhu3, Ashaq Hussain Bhat3, Arunsaikiran Senthilkumar3, Raj Pranap Arun1, Sharath Asokan4, Sachin S. Gunthe2 & Rama S. Verma1,5* Dental caries is the most prevalent oral disease afecting nearly 70% of children in India and elsewhere. Micro-ecological niche based acidifcation due to dysbiosis in oral microbiome are crucial for caries onset and progression. Here we report the tooth bacteriome diversity compared in Indian children with caries free (CF), severe early childhood caries (SC) and recurrent caries (RC). High quality V3–V4 amplicon sequencing revealed that SC exhibited high bacterial diversity with unique combination and interrelationship. Gracillibacteria_GN02 and TM7 were unique in CF and SC respectively, while Bacteroidetes, Fusobacteria were signifcantly high in RC. Interestingly, we found Streptococcus oralis subsp. tigurinus clade 071 in all groups with signifcant abundance in SC and RC. Positive correlation between low and high abundant bacteria as well as with TCS, PTS and ABC transporters were seen from co-occurrence network analysis. This could lead to persistence of SC niche resulting in RC. Comparative in vitro assessment of bioflm formation showed that the standard culture of S. oralis and its phylogenetically similar clinical isolates showed profound bioflm formation and augmented the growth and enhanced bioflm formation in S. mutans in both dual and multispecies cultures. Interaction among more than 700 species of microbiota under diferent micro-ecological niches of the human oral cavity1,2 acts as a primary defense against various pathogens. Tis has been observed to play a signifcant role in child’s oral and general health.