Shweta Agarwal Dr. Ranjana Mehrotra
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
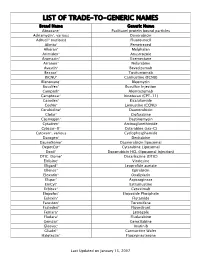
Trade-To-Generic Names
LIST OF TRADE-TO-GENERIC NAMES Brand Name Generic Name Abraxane® Paclitaxel protein bound particles Adriamycin®, various Doxorubicin Adrucil® (various) Fluorouracil Alimta® Pemetrexed Alkeran® Melphalan Arimidex® Anastrozole Aromasin® Exemestane Arranon® Nelarabine Avastin® Bevacizumab Bexxar® Tositumomab BiCNU® Carmustine (BCNU) Blenoxane® Bleomycin Busulfex® Busulfan Injection Campath® Alemtuzumab Camptosar® Irinotecan (CPT-11) Casodex® Bicalutamide CeeNu® Lomustine (CCNU) Cerubidine® Daunorubicin Clolar® Clofarabine Cosmegen® Dactinomycin Cytadren® Aminoglutethimide Cytosar-U® Cytarabine (ara-C) Cytoxan®, various Cyclophosphamide Dacogen® Decitabine DaunoXome® Daunorubicin liposomal DepotCyt® Cytarabine Liposomal Doxil® Doxorubicin HCL (liposomal injection) DTIC-Dome® Dacarbazine (DTIC) Eldisine® Vindesine Eligard® Leuprolide acetate Ellence® Epirubicin Eloxatin® Oxaliplatin Elspar® Asparaginase EmCyt® Estramustine Erbitux® Cetuximab Etopofos® Etoposide Phosphate Eulexin® Flutamide Fareston® Toremifene Faslodex® Fluvestrant Femara® Letrozole Fludara® Fludarabine Gemzar® Gemcitabine Gleevec® Imatinib Gliadel® Carmustine Wafer Halotestin® Fluoxymesterone Last Updated on January 15, 2007 Brand Name Generic Name Herceptin® Trastuzumab Hexalen® Altretamine Hycamtin® Topotecan Hydrea® Hydroxyurea Idamycin® Idarubicin Ifex® Ifosfamide Intron A® Interferon alfa-2b Iressa® Gefitinib Leukeran® Chlorambucil Leukine® Sargramostim Leustatin® Cladribine Lupron depot® Leuprolide acetate depot Lupron® Leuprolide acetate Matulane® Procarbazine Megace® -

Phenotype Microarrays Panels PM-M1 to PM-M14
Phenotype MicroArrays™ Panels PM-M1 to PM-M14 for Phenotypic Characterization of Mammalian Cells Assays: Energy Metabolism Pathways Ion and Hormone Effects on Cells Sensitivity to Anti-Cancer Agents and for Optimizing Culture Conditions for Mammalian Cells PRODUCT DESCRIPTIONS AND INSTRUCTIONS FOR USE PM-M1 Cat. #13101 PM-M2 Cat. #13102 PM-M3 Cat. #13103 PM-M4 Cat. #13104 PM-M5 Cat. #13105 PM-M6 Cat. #13106 PM-M7 Cat. #13107 PM-M8 Cat. #13108 PM-M11 Cat. #13111 PM-M12 Cat. #13112 PM-M13 Cat. #13113 PM-M14 Cat. #13114 © 2016 Biolog, Inc. All rights reserved Printed in the United States of America 00P 134 Rev F February 2020 - 1 - CONTENTS I. Introduction ...................................................................................................... 2 a. Overview ................................................................................................... 2 b. Background ............................................................................................... 2 c. Uses ........................................................................................................... 2 d. Advantages ................................................................................................ 3 II. Product Description, PM-M1 to M4 ................................................................ 3 III. Protocols, PM-M1 to M4 ................................................................................. 7 a. Materials Required .................................................................................... 7 b. Determination -

NINDS Custom Collection II
ACACETIN ACEBUTOLOL HYDROCHLORIDE ACECLIDINE HYDROCHLORIDE ACEMETACIN ACETAMINOPHEN ACETAMINOSALOL ACETANILIDE ACETARSOL ACETAZOLAMIDE ACETOHYDROXAMIC ACID ACETRIAZOIC ACID ACETYL TYROSINE ETHYL ESTER ACETYLCARNITINE ACETYLCHOLINE ACETYLCYSTEINE ACETYLGLUCOSAMINE ACETYLGLUTAMIC ACID ACETYL-L-LEUCINE ACETYLPHENYLALANINE ACETYLSEROTONIN ACETYLTRYPTOPHAN ACEXAMIC ACID ACIVICIN ACLACINOMYCIN A1 ACONITINE ACRIFLAVINIUM HYDROCHLORIDE ACRISORCIN ACTINONIN ACYCLOVIR ADENOSINE PHOSPHATE ADENOSINE ADRENALINE BITARTRATE AESCULIN AJMALINE AKLAVINE HYDROCHLORIDE ALANYL-dl-LEUCINE ALANYL-dl-PHENYLALANINE ALAPROCLATE ALBENDAZOLE ALBUTEROL ALEXIDINE HYDROCHLORIDE ALLANTOIN ALLOPURINOL ALMOTRIPTAN ALOIN ALPRENOLOL ALTRETAMINE ALVERINE CITRATE AMANTADINE HYDROCHLORIDE AMBROXOL HYDROCHLORIDE AMCINONIDE AMIKACIN SULFATE AMILORIDE HYDROCHLORIDE 3-AMINOBENZAMIDE gamma-AMINOBUTYRIC ACID AMINOCAPROIC ACID N- (2-AMINOETHYL)-4-CHLOROBENZAMIDE (RO-16-6491) AMINOGLUTETHIMIDE AMINOHIPPURIC ACID AMINOHYDROXYBUTYRIC ACID AMINOLEVULINIC ACID HYDROCHLORIDE AMINOPHENAZONE 3-AMINOPROPANESULPHONIC ACID AMINOPYRIDINE 9-AMINO-1,2,3,4-TETRAHYDROACRIDINE HYDROCHLORIDE AMINOTHIAZOLE AMIODARONE HYDROCHLORIDE AMIPRILOSE AMITRIPTYLINE HYDROCHLORIDE AMLODIPINE BESYLATE AMODIAQUINE DIHYDROCHLORIDE AMOXEPINE AMOXICILLIN AMPICILLIN SODIUM AMPROLIUM AMRINONE AMYGDALIN ANABASAMINE HYDROCHLORIDE ANABASINE HYDROCHLORIDE ANCITABINE HYDROCHLORIDE ANDROSTERONE SODIUM SULFATE ANIRACETAM ANISINDIONE ANISODAMINE ANISOMYCIN ANTAZOLINE PHOSPHATE ANTHRALIN ANTIMYCIN A (A1 shown) ANTIPYRINE APHYLLIC -

A Comparison Between Triplet and Doublet Chemotherapy in Improving
Guo et al. BMC Cancer (2019) 19:1125 https://doi.org/10.1186/s12885-019-6294-9 RESEARCH ARTICLE Open Access A comparison between triplet and doublet chemotherapy in improving the survival of patients with advanced gastric cancer: a systematic review and meta-analysis Xinjian Guo1, Fuxing Zhao1, Xinfu Ma1, Guoshuang Shen1, Dengfeng Ren1, Fangchao Zheng1,2,3, Feng Du4, Ziyi Wang1, Raees Ahmad1, Xinyue Yuan1, Junhui Zhao1* and Jiuda Zhao1* Abstract Background: Chemotherapy can improve the survival of patients with advanced gastric cancer. However, whether triplet chemotherapy can further improve the survival of patients with advanced gastric cancer compared with doublet chemotherapy remains controversial. This study reviewed and updated all published and eligible randomized controlled trials (RCTs) to compare the efficacy, prognosis, and toxicity of triplet chemotherapy with doublet chemotherapy in patients with advanced gastric cancer. Methods: RCTs on first-line chemotherapy in advanced gastric cancer on PubMed, Embase, and the Cochrane Register of Controlled Trials and all abstracts from the annual meetings of the European Society for Medical Oncology (ESMO) and the American Society of Clinical Oncology conferences up to October 2018 were searched. The primary outcome was overall survival, while the secondary outcomes were progression-free survival (PFS), time to progress (TTP), objective response rate (ORR), and toxicity. Results: Our analysis included 23 RCTs involving 4540 patients and 8 types of triplet and doublet chemotherapy regimens, and systematic review and meta-analysis revealed that triplet chemotherapy was superior compared with doublet chemotherapy in terms of improving median OS (HR = 0.92; 95% CI, 0.86–0.98; P = 0.02) and PFS (HR = 0.82; 95% CI, 0.69–0.97; P = 0.02) and TTP (HR = 0.92; 95% CI, 0.86–0.98; P = 0.02) and ORR (OR = 1.21; 95% CI, 1.12–1.31; P < 0.0001) among overall populations. -

Covalently-Linked Complexes and Methods for Enhanced Cytotoxicity and Imaging
Europaisches Patentamt 19 European Patent Office Office europeen des brevets © Publication number: 0 359 347 B1 12 EUROPEAN PATENT SPECIFICATION © Date of publication of patent specification © int. ci.5: A61K 47/42, A61K 49/02, 23.12.92 Bulletin 92/52 A61K 43/00 © Application number : 89250014.1 © Date of filing : 14.08.89 @) Covalently-linked complexes and methods for enhanced cytotoxicity and imaging. © Priority : 15.08.88 US 232337 © Proprietor : NEORX CORPORATION 410 West Harrison Street Seattle Washington 98119 (US) @ Date of publication of application 21.03.90 Bulletin 90/12 © Inventor : Anderson, David C. 2415 Thorndyke Avenue No.301 © Publication of the grant of the patent : Seattle Washington 98199 (US) 23.12.92 Bulletin 92/52 Inventor : Morgan, A. Charles 803 Driftwood Place Edmonds Washington 98020 (US) @ Designated Contracting States : Inventor : Abrams, Paul G. AT BE CH DE ES FR GB GR IT LI LU NL SE 2125 First Avenue No. 1602 Seattle Washington 98121 (US) Inventor : Nichols, Everett J. © References cited : 12525 17th Avenue N.E. EP-A- 0 282 057 Seattle Washington 98125 (US) CHEMICAL ABSTRACTS, vol. 108, no. 7, 1987, Inventor : Fritzberg, Alan R. abstract no. 48913a, Columbus, Ohio, US; J.M. 16703 74th Place West BOGGS et al.: Edmonds Washington 98020 (US) "Antigen-targetedliposome-encapsulated methotrexate specifically kills lymphocytes sensitized to the nonapeptide of myelin basic © Representative : Wablat, Wolfgang, Dr. Dr. protein ", & J.NEUROIMMUNOL, 17(1), 35-48 Potsdamer Chaussee 48 W-1000 Berlin 38 (DE) CO h- CO If) CO Note : Within nine months from the publication of the mention of the grant of the European patent, any person may give notice to the European Patent Office of opposition to the European patent granted. -

Appendix C Medication Tables
Appendix C Medication Tables Note: The medication tables are not meant to be inclusive lists of all available therapeutic agents. Approved medication tables will be updated regularly. Discrepancies must be reported. See Resource Section of this manual for additional contact information. Release Notes: Aspirin Table Version 1.0 Table 1.1 Aspirin and Aspirin-Containing Medications Acetylsalicylic Acid Acuprin 81 Alka-Seltzer Alka-Seltzer Morning Relief Anacin Arthritis Foundation Aspirin Arthritis Pain Ascriptin Arthritis Pain Formula ASA ASA Baby ASA Baby Chewable ASA Baby Coated ASA Bayer ASA Bayer Children's ASA Buffered ASA Children's ASA EC ASA Enteric Coated ASA/Maalox Ascriptin Aspergum Aspir-10 Aspir-Low Aspir-Lox Aspir-Mox Aspir-Trin Aspirbuf Aspircaf Aspirin Aspirin Baby Aspirin Bayer Aspirin Bayer Children's Aspirin Buffered Aspirin Child Aspirin Child Chewable Aspirin Children's Aspirin EC Aspirin Enteric Coated Specifications Manual for National Appendix C-1 Hospital Quality Measures Table 1.1 Aspirin and Aspirin-Containing Medications (continued) Aspirin Litecoat Aspirin Lo-Dose Aspirin Low Strength Aspirin Tri-Buffered Aspirin, Extended Release Aspirin/butalbital/caffeine Aspirin/caffeine Aspirin/pravachol Aspirin/pravastatin Aspirtab Bayer Aspirin Bayer Aspirin PM Extra Strength Bayer Children’s Bayer EC Bayer Enteric Coated Bayer Low Strength Bayer Plus Buffered ASA Buffered Aspirin Buffered Baby ASA Bufferin Bufferin Arthritis Strength Bufferin Extra Strength Buffex Cama Arthritis Reliever Child’s Aspirin Coated Aspirin -

(12) United States Patent (10) Patent No.: US 9,283,215 B2 Zeldis (45) Date of Patent: *Mar
US009283215 B2 (12) United States Patent (10) Patent No.: US 9,283,215 B2 Zeldis (45) Date of Patent: *Mar. 15, 2016 (54) METHODS FORTREATING MULTIPLE (2013.01); A61 K3I/4439 (2013.01); A61 K MYELOMAUSING 4-(AMINO)-2- 31/475 (2013.01); A61 K3I/515 (2013.01); (2,6-DIOXO(3-PIPERIDYL))-ISOINDOLINE- A6 IK3I/573 (2013.01); A61 K3I/675 SES COMBINATION WITH (2013.01); A61 K3I/704 (2013.01); A61 K 31/7048 (2013.01); A61 K35/12 (2013.01); (71) Applicant: Celgene Corporation, Summit, NJ (US) A61K39/00II (2013.01); A61 K39/3955 (2013.01); A61K 45/06 (2013.01); A61 K (72) Inventor: Jerome B. Zeldis, Princeton, NJ (US) 2300/00 (2013.01) (58) Field of Classification Search (73) Assignee: Celgene Corporation, Summit, NJ (US) CPC ........... A61K 31/4462: A61K 31/4035; A61 K (*) Notice: Subject to any disclaimer, the term of this 31f445 patent is extended or adjusted under 35 USPC .......................................... 514/323,329–330 U.S.C. 154(b) by 0 days. See application file for complete search history. This patent is Subject to a terminal dis claimer. (56) References Cited (21) Appl. No.: 14/201,069 U.S. PATENT DOCUMENTS (22) Filed: Mar. 7, 2014 3.536.809 A 10, 1970 Applezweig 3,598,123 A 8, 1971 Zaffaroni et al. O O 3,845,770 A 11/1974 Theeuwes et al. (65) Prior Publication Data 3,916,899 A 1 1/1975 Theeuwes et al. US 2014/0186404 A1 Jul. 3, 2014 4,008,719 A 2f1977 Theeuwes et al. 4,810,643 A 3, 1989 Souza Related U.S. -

Antigen Binding Protein and Its Use As Addressing Product for the Treatment of Cancer
(19) TZZ 58Z9A_T (11) EP 2 589 609 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.05.2013 Bulletin 2013/19 C07K 16/28 (2006.01) (21) Application number: 11306416.6 (22) Date of filing: 03.11.2011 (84) Designated Contracting States: (72) Inventors: AL AT BE BG CH CY CZ DE DK EE ES FI FR GB •Beau-Larvor, Charlotte GR HR HU IE IS IT LI LT LU LV MC MK MT NL NO 74520 Jonzier Epagny (FR) PL PT RO RS SE SI SK SM TR • Goetsch, Liliane Designated Extension States: 74130 Ayze (FR) BA ME (74) Representative: Regimbeau (83) Declaration under Rule 32(1) EPC (expert 20, rue de Chazelles solution) 75847 Paris Cedex 17 (FR) (71) Applicant: PIERRE FABRE MEDICAMENT 92100 Boulogne-Billancourt (FR) (54) Antigen binding protein and its use as addressing product for the treatment of cancer (57) The present invention relates to an antigen bind- of Axl, being internalized into the cell. The invention also ing protein, in particular a monoclonal antibody, capable comprises the use of said antigen binding protein as an of binding specifically to the protein Axl as well as the addressing product in conjugation with other anti- cancer amino and nucleic acid sequences coding for said pro- compounds,such as toxins, radio- elements ordrugs, and tein. From one aspect, the invention relates to an antigen the use of same for the treatment of certain cancers. binding protein, or antigen binding fragments, capable of binding specifically to Axl and, by inducing internalization EP 2 589 609 A1 Printed by Jouve, 75001 PARIS (FR) EP 2 589 609 A1 Description [0001] The present invention relates to a novel antigen binding protein, in particular a monoclonal antibody, capable of binding specifically to the protein Axl as well as the amino and nucleic acid sequences coding for said protein. -

Combined Modality Approaches in the Management of Adult Glioblastoma
REVIEW ARTICLE published: 28 October 2011 doi: 10.3389/fonc.2011.00036 Combined modality approaches in the management of adult glioblastoma Haider A. Shirazi 1, Sean Grimm2, Jeffrey Raizer 2 and Minesh P.Mehta1* 1 Department of Radiation Oncology, Feinberg School of Medicine, Northwestern University, Chicago, IL, USA 2 Department of Neurology, Feinberg School of Medicine, Northwestern University, Chicago, IL, USA Edited by: Over the past two decades, management of newly diagnosed glioblastoma has undergone Jann Sarkaria, Mayo Clinic, USA significant evolution. While surgery has long been a mainstay of management for this dis- Reviewed by: ease, and while radiotherapy has a proven survival role, initial efforts at radiotherapy dose Elizabeth Yan, Mayo Clinic, USA Daniel Ma, Mayo Clinic, USA escalation, use of radiosurgery, brachytherapy, and altered fractionation did not improve *Correspondence: patient survival. Recently, multiple modality therapy integrating maximal safe resection, Minesh P.Mehta, Department of postoperative radiation, and new systemic therapies have resulted in improved patient out- Radiation Oncology, Feinberg School comes compared with older regimens utilizing surgery and postoperative radiation alone. of Medicine, Northwestern Numerous trials are currently underway investigating the combination of surgery, radiation, University, 251 E. Huron Street, LC-178, Chicago, IL 60611, USA. and systemic therapy with targeted agents to find ways to further improve outcomes for e-mail: [email protected] adults with glioblastoma. Keywords: glioblastoma, radiotherapy, chemotherapy INTRODUCTION earlier, there are insufficient level 1 data to conclude that complete Glioblastoma (GBM) remains a highly lethal and aggressive tumor resection imparts a survival benefit; however, in a large single insti- with dismal prognosis. -

(12) Patent Application Publication (10) Pub. No.: US 2015/0150995 A1 Taft, III Et Al
US 2015O150995A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2015/0150995 A1 Taft, III et al. (43) Pub. Date: Jun. 4, 2015 (54) CONJUGATED ANTI-MICROBIAL Publication Classification COMPOUNDS AND CONUGATED ANT-CANCER COMPOUNDS AND USES (51) Int. Cl. THEREOF A647/48 (2006.01) A63/546 (2006.01) (71) Applicant: PONO CORPORATION, Honolulu, HI A633/38 (2006.01) (US) (52) U.S. Cl. CPC ........... A61K47/480.15 (2013.01); A61K33/38 (72) Inventors: Karl Milton Taft, III, Honolulu, HI (2013.01); A61 K3I/546 (2013.01) (US); Jarred Roy Engelking, Honolulu, HI (US) (57) ABSTRACT (73) Assignee: PONO CORPORATION, Honolulu, HI Disclosed herein are synthesis methods for generation of (US) conjugated anti-microbial compounds and conjugated anti cancer compounds. Several embodiments, related to the uses (21) Appl.ppl. NNo.: 14/418,9079 of Such compoundsp in the treatment of infections, in particu lar those caused by drug-resistant bacteria. Some embodi (22) PCT Filed: Aug. 9, 2013 ments relate to targeting cancer based on the metabolic sig (86). PCT No.: PCT/US2O13/O54391 nature of tumor cells. S371 (c)(1), (2) Date: Jan. 30, 2015 NH Related U.S. Application Data O Ag" (60) Provisional application No. 61/742,443, filed on Aug. B-Lactam Silver Ion 9, 2012, provisional application No. 61/742,444, filed on Aug. 9, 2012. O O O O O O HSONaNO -pE (CHO)2SO2 OEt Br2 OEt 2W4 OH NOMe 1 2 3 Q Q H.N.S NH, NHT chicci -VV653C(CH) NaOH Bra-oe 2 2 S1N (C6H5)3C- SNN a NOMe MeO -co. -

Stembook 2018.Pdf
The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances FORMER DOCUMENT NUMBER: WHO/PHARM S/NOM 15 WHO/EMP/RHT/TSN/2018.1 © World Health Organization 2018 Some rights reserved. This work is available under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 IGO licence (CC BY-NC-SA 3.0 IGO; https://creativecommons.org/licenses/by-nc-sa/3.0/igo). Under the terms of this licence, you may copy, redistribute and adapt the work for non-commercial purposes, provided the work is appropriately cited, as indicated below. In any use of this work, there should be no suggestion that WHO endorses any specific organization, products or services. The use of the WHO logo is not permitted. If you adapt the work, then you must license your work under the same or equivalent Creative Commons licence. If you create a translation of this work, you should add the following disclaimer along with the suggested citation: “This translation was not created by the World Health Organization (WHO). WHO is not responsible for the content or accuracy of this translation. The original English edition shall be the binding and authentic edition”. Any mediation relating to disputes arising under the licence shall be conducted in accordance with the mediation rules of the World Intellectual Property Organization. Suggested citation. The use of stems in the selection of International Nonproprietary Names (INN) for pharmaceutical substances. Geneva: World Health Organization; 2018 (WHO/EMP/RHT/TSN/2018.1). Licence: CC BY-NC-SA 3.0 IGO. Cataloguing-in-Publication (CIP) data. -
![Download This Topic [PDF]](https://docslib.b-cdn.net/cover/6342/download-this-topic-pdf-3566342.webp)
Download This Topic [PDF]
cancer.org | 1.800.227.2345 Known and Probable Human Carcinogens In general, the American Cancer Society does not determine if something causes cancer (that is, if it is a carcinogen). Instead, we rely on the determinations of other respected agencies, such as the International Agency for Research on Cancer (IARC) and the US National Toxicology Program (NTP). The lists below are from IARC and NTP, and more information on each of these known and probable human carcinogens can be found on their websites. To learn more about these agencies and how they study and classify cancer causes, see Determining if Something Is a Carcinogen1. What you should know ● The IARC and NTP act independently. Many known or suspected carcinogens appear on both organization’s lists; however, if a substance or exposure is only on one agency’s list, this it does not necessarily mean there is a controversy, as one agency may not have evaluated it. ● These lists are alphabetical, but many of the substances and exposures here can go by different names. This can make it hard to find a particular substance on one or both of these lists. ● These lists include only those agents that have been evaluated by the agencies. These agencies tend to focus on substances and exposures most likely to cause cancer, but there are many others that have not been fully studied yet. ● These lists include agents that have been classified as known and probable human carcinogens. The lists do not include substances that have been classified as possible carcinogens, for which the evidence is not as strong.