Acetyl Group Coordinated Progression Through the Catalytic Cycle of an Arylalkylamine N-Acetyltransferase
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Table S1
Entrez Gene Symbol Gene Name Affymetrix EST Glomchip SAGE Stanford Literature HPA confirmed Gene ID Profiling profiling Profiling Profiling array profiling confirmed 1 2 A2M alpha-2-macroglobulin 0 0 0 1 0 2 10347 ABCA7 ATP-binding cassette, sub-family A (ABC1), member 7 1 0 0 0 0 3 10350 ABCA9 ATP-binding cassette, sub-family A (ABC1), member 9 1 0 0 0 0 4 10057 ABCC5 ATP-binding cassette, sub-family C (CFTR/MRP), member 5 1 0 0 0 0 5 10060 ABCC9 ATP-binding cassette, sub-family C (CFTR/MRP), member 9 1 0 0 0 0 6 79575 ABHD8 abhydrolase domain containing 8 1 0 0 0 0 7 51225 ABI3 ABI gene family, member 3 1 0 1 0 0 8 29 ABR active BCR-related gene 1 0 0 0 0 9 25841 ABTB2 ankyrin repeat and BTB (POZ) domain containing 2 1 0 1 0 0 10 30 ACAA1 acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiol 0 1 0 0 0 11 43 ACHE acetylcholinesterase (Yt blood group) 1 0 0 0 0 12 58 ACTA1 actin, alpha 1, skeletal muscle 0 1 0 0 0 13 60 ACTB actin, beta 01000 1 14 71 ACTG1 actin, gamma 1 0 1 0 0 0 15 81 ACTN4 actinin, alpha 4 0 0 1 1 1 10700177 16 10096 ACTR3 ARP3 actin-related protein 3 homolog (yeast) 0 1 0 0 0 17 94 ACVRL1 activin A receptor type II-like 1 1 0 1 0 0 18 8038 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 1 0 0 0 0 19 8751 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 1 0 0 0 0 20 8728 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 1 0 0 0 0 21 81792 ADAMTS12 ADAM metallopeptidase with thrombospondin type 1 motif, 12 1 0 0 0 0 22 9507 ADAMTS4 ADAM metallopeptidase with thrombospondin type 1 -
Index Mutated* Normal* FC P Value FDR** Probe Set Description Gene Symbol
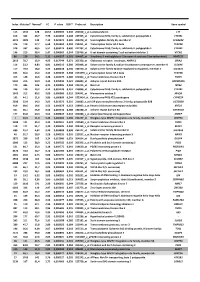
Index Mutated* Normal* FC P value FDR** Probe set Description Gene symbol 113 1342 128 10,52 0,000503 0,240 202018_s_at Lactotransferrin LTF 616 362 46,7 7,76 0,003493 0,308 237395_at Cytochrome P450, family 4, subfamily Z, polypeptide 1 CYP4Z1 3073 1009 142 7,10 0,021674 0,385 206378_at Secretoglobin, family 2A, member 2 SCGB2A2 376 119 17,7 6,68 0,001962 0,284 214451_at Transcription factor AP-2 beta TFAP2B 578 337 60,5 5,57 0,003174 0,300 227702_at Cytochrome P450, family 4, subfamily X, polypeptide 1 CYP4X1 146 210 38,4 5,47 0,000669 0,244 219768_at V-set domain containing T cell activation inhibitor 1 VTCN1 86 129 24,2 5,31 0,000327 0,201 204607_at 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (mitochondrial) HMGCS2 2613 78,7 15,9 4,95 0,017744 0,371 205358_at Glutamate receptor, ionotropic, AMPA 2 GRIA2 116 23,2 4,83 4,81 0,000513 0,240 203908_at Solute carrier family 4, sodium bicarbonate cotransporter, member 4 SLC4A4 117 79,4 18,0 4,42 0,000518 0,240 239723_at Solute carrier family 40 (iron-regulated transporter), member 1 SLC40A1 625 56,6 13,0 4,34 0,003549 0,308 1553394_a_atTranscription factor AP-2 beta TFAP2B 413 148 34,6 4,28 0,002175 0,286 240304_s_at Transmembrane channel-like 5 TMC5 5181 216 50,9 4,25 0,039946 0,421 223864_at Ankyrin repeat domain 30A ANKRD30A 179 446 106 4,21 0,000826 0,244 223315_at Netrin 4 NTN4 246 120 29,2 4,12 0,001128 0,251 210096_at Cytochrome P450, family 4, subfamily B, polypeptide 1 CYP4B1 1043 312 80,2 3,89 0,006080 0,319 204041_at Monoamine oxidase B MAOB 192 44,1 11,6 3,80 0,000899 0,244 -

Appendix 2. Significantly Differentially Regulated Genes in Term Compared with Second Trimester Amniotic Fluid Supernatant
Appendix 2. Significantly Differentially Regulated Genes in Term Compared With Second Trimester Amniotic Fluid Supernatant Fold Change in term vs second trimester Amniotic Affymetrix Duplicate Fluid Probe ID probes Symbol Entrez Gene Name 1019.9 217059_at D MUC7 mucin 7, secreted 424.5 211735_x_at D SFTPC surfactant protein C 416.2 206835_at STATH statherin 363.4 214387_x_at D SFTPC surfactant protein C 295.5 205982_x_at D SFTPC surfactant protein C 288.7 1553454_at RPTN repetin solute carrier family 34 (sodium 251.3 204124_at SLC34A2 phosphate), member 2 238.9 206786_at HTN3 histatin 3 161.5 220191_at GKN1 gastrokine 1 152.7 223678_s_at D SFTPA2 surfactant protein A2 130.9 207430_s_at D MSMB microseminoprotein, beta- 99.0 214199_at SFTPD surfactant protein D major histocompatibility complex, class II, 96.5 210982_s_at D HLA-DRA DR alpha 96.5 221133_s_at D CLDN18 claudin 18 94.4 238222_at GKN2 gastrokine 2 93.7 1557961_s_at D LOC100127983 uncharacterized LOC100127983 93.1 229584_at LRRK2 leucine-rich repeat kinase 2 HOXD cluster antisense RNA 1 (non- 88.6 242042_s_at D HOXD-AS1 protein coding) 86.0 205569_at LAMP3 lysosomal-associated membrane protein 3 85.4 232698_at BPIFB2 BPI fold containing family B, member 2 84.4 205979_at SCGB2A1 secretoglobin, family 2A, member 1 84.3 230469_at RTKN2 rhotekin 2 82.2 204130_at HSD11B2 hydroxysteroid (11-beta) dehydrogenase 2 81.9 222242_s_at KLK5 kallikrein-related peptidase 5 77.0 237281_at AKAP14 A kinase (PRKA) anchor protein 14 76.7 1553602_at MUCL1 mucin-like 1 76.3 216359_at D MUC7 mucin 7, -

Whole Egg Consumption Increases Gene Expression Within the Glutathione Pathway in the Liver of Zucker Diabetic Fatty Rats
Food Science and Human Nutrition Publications Food Science and Human Nutrition 11-3-2020 Whole egg consumption increases gene expression within the glutathione pathway in the liver of Zucker Diabetic Fatty rats Joe L. Webb Iowa State University Amanda E. Bries Iowa State University, [email protected] Brooke Vogel Iowa State University Claudia Carrillo Iowa State University, [email protected] Lily Harvison Iowa State University, [email protected] See next page for additional authors Follow this and additional works at: https://lib.dr.iastate.edu/fshn_hs_pubs Part of the Dietetics and Clinical Nutrition Commons, Endocrinology, Diabetes, and Metabolism Commons, Exercise Science Commons, Food Chemistry Commons, Human and Clinical Nutrition Commons, and the Molecular, Genetic, and Biochemical Nutrition Commons The complete bibliographic information for this item can be found at https://lib.dr.iastate.edu/ fshn_hs_pubs/38. For information on how to cite this item, please visit http://lib.dr.iastate.edu/ howtocite.html. This Article is brought to you for free and open access by the Food Science and Human Nutrition at Iowa State University Digital Repository. It has been accepted for inclusion in Food Science and Human Nutrition Publications by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Whole egg consumption increases gene expression within the glutathione pathway in the liver of Zucker Diabetic Fatty rats Abstract Nutrigenomic evidence supports the idea that Type 2 Diabetes Mellitus (T2DM) arises due to the interactions between the transcriptome, individual genetic profiles, lifestyle, and diet. Since eggs are a nutrient dense food containing bioactive ingredients that modify gene expression, our goal was to examine the role of whole egg consumption on the transcriptome during T2DM. -

Supplemental Table S1. Primers for Sybrgreen Quantitative RT-PCR Assays
Supplemental Table S1. Primers for SYBRGreen quantitative RT-PCR assays. Gene Accession Primer Sequence Length Start Stop Tm GC% GAPDH NM_002046.3 GAPDH F TCCTGTTCGACAGTCAGCCGCA 22 39 60 60.43 59.09 GAPDH R GCGCCCAATACGACCAAATCCGT 23 150 128 60.12 56.52 Exon junction 131/132 (reverse primer) on template NM_002046.3 DNAH6 NM_001370.1 DNAH6 F GGGCCTGGTGCTGCTTTGATGA 22 4690 4711 59.66 59.09% DNAH6 R TAGAGAGCTTTGCCGCTTTGGCG 23 4797 4775 60.06 56.52% Exon junction 4790/4791 (reverse primer) on template NM_001370.1 DNAH7 NM_018897.2 DNAH7 F TGCTGCATGAGCGGGCGATTA 21 9973 9993 59.25 57.14% DNAH7 R AGGAAGCCATGTACAAAGGTTGGCA 25 10073 10049 58.85 48.00% Exon junction 9989/9990 (forward primer) on template NM_018897.2 DNAI1 NM_012144.2 DNAI1 F AACAGATGTGCCTGCAGCTGGG 22 673 694 59.67 59.09 DNAI1 R TCTCGATCCCGGACAGGGTTGT 22 822 801 59.07 59.09 Exon junction 814/815 (reverse primer) on template NM_012144.2 RPGRIP1L NM_015272.2 RPGRIP1L F TCCCAAGGTTTCACAAGAAGGCAGT 25 3118 3142 58.5 48.00% RPGRIP1L R TGCCAAGCTTTGTTCTGCAAGCTGA 25 3238 3214 60.06 48.00% Exon junction 3124/3125 (forward primer) on template NM_015272.2 Supplemental Table S2. Transcripts that differentiate IPF/UIP from controls at 5%FDR Fold- p-value Change Transcript Gene p-value p-value p-value (IPF/UIP (IPF/UIP Cluster ID RefSeq Symbol gene_assignment (Age) (Gender) (Smoking) vs. C) vs. C) NM_001178008 // CBS // cystathionine-beta- 8070632 NM_001178008 CBS synthase // 21q22.3 // 875 /// NM_0000 0.456642 0.314761 0.418564 4.83E-36 -2.23 NM_003013 // SFRP2 // secreted frizzled- 8103254 NM_003013 -

Supplementary Table 1A. Genes Significantly Altered in A4573 ESFT
Supplementary Table 1A. Genes significantly altered in A4573 ESFT cells following BMI-1knockdown genesymbol genedescription siControl siBMI1 FC Direction P-value AASS aminoadipate-semialdehyde synthase | tetra-peptide repeat homeobox-like6.68 7.24 1.5 Up 0.007 ABCA2 ATP-binding cassette, sub-family A (ABC1), member 2 | neural5.44 proliferation,6.3 differentiation1.8 and Upcontrol, 1 0.006 ABHD4 abhydrolase domain containing 4 7.51 6.69 1.8 Down 0.002 ACACA acetyl-Coenzyme A carboxylase alpha | peroxiredoxin 5 | similar6.2 to High mobility7.26 group2.1 protein UpB1 (High mobility0.009 group protein 1) (HMG-1) (Amphoterin) (Heparin-binding protein p30) | Coenzyme A synthase ACAD9 acyl-Coenzyme A dehydrogenase family, member 9 9.25 8.59 1.6 Down 0.008 ACBD3 acyl-Coenzyme A binding domain containing 3 7.89 8.53 1.6 Up 0.008 ACCN2 amiloride-sensitive cation channel 2, neuronal 5.47 6.28 1.8 Up 0.005 ACIN1 apoptotic chromatin condensation inducer 1 7.15 7.79 1.6 Up 0.008 ACPL2 acid phosphatase-like 2 6.04 7.6 2.9 Up 0.000 ACSL4 acyl-CoA synthetase long-chain family member 4 6.72 5.8 1.9 Down 0.001 ACTA2 actin, alpha 2, smooth muscle, aorta 9.18 8.44 1.7 Down 0.003 ACYP1 acylphosphatase 1, erythrocyte (common) type 7.09 7.66 1.5 Up 0.009 ADA adenosine deaminase 6.34 7.1 1.7 Up 0.009 ADAL adenosine deaminase-like 7.88 6.89 2.0 Down 0.006 ADAMTS1 ADAM metallopeptidase with thrombospondin type 1 motif, 1 6.57 7.65 2.1 Up 0.000 ADARB1 adenosine deaminase, RNA-specific, B1 (RED1 homolog rat) 6.49 7.13 1.6 Up 0.008 ADCY9 adenylate cyclase 9 6.5 7.18 -

Supplemental Figure 1 Characterization of Lung Tumors In
Supplemental Figure 1 Characterization of lung tumors in KC mice. (A) Scatter plots of visible spontaneous tumors counted on the lung surfaces at necropsy. Each dot represents a value from a single mouse, and the bars and whiskers show the mean SD for each cohort from a single experiment. P value, two-tailed Student’s t-test. (B) Histologic analysis of spontaneous lung tumors in (A). Results indicate the numbers of lung lesions per histologic subtype per genotype. N, cohort sample sizes. P values, Fisher's exact test. Supplemental Figure 2 Surfactant protein C (SPC) expression in p21-deficient (KC) and –replete (K) tumor cell lines. Immunofluorescence staining with anti-surfactant protein C antibody (green) or normal IgG as a negative control. DAPI, blue. SPC/DAPI and IgG/DAPI are merged images. Scale bar, 20 µm. Supplemental Figure 3 Somatic recombination of latent KrasG12D allele and detection of p21 protein in p21-deficient (KC) and –replete (K) tumor cell lines. (A) PCR analysis of the K-rasLA1 and K-rasWT alleles in genomic DNA. Somatic recombination of the K-rasLA1 allele leads to loss of the PCR product (upper band, arrow). Wild-type K-ras allele detected (250 bp). Tail DNAs from KrasLA1 mice (KrasLA1/+) and wild-type mice (Kras+/+) were used as positive and negative controls, respectively, for the presence of the germline K-rasLA1 allele. (B) Immunoblot analysis of p21 expression. Actin was used as a loading control. Supplemental Figure 4 Growth and metastasis of p21-deficient (KC) tumor cell lines in Cdkn1a-null or –wild-type mice. -

Directed Differentiation of Human Embryonic Stem Cells Into Haematopoietic and Definitive Endodermal Lineages
DIRECTED DIFFERENTIATION OF HUMAN EMBRYONIC STEM CELLS INTO HAEMATOPOIETIC AND DEFINITIVE ENDODERMAL LINEAGES ABRAHAM SUMAN MARY (M.Sc MICROBIOLOGY, UNIV. OF MUMBAI, INDIA) A THESIS SUBMITTED FOR THE DEGREE OF MASTER OF SCIENCE DEPARTMENT OF BIOCHEMISTRY NATIONAL UNIVERSITY OF SINGAPORE 2009 ACKNOWLEDGEMENTS In all things I give YOU glory! You have always led me through amazing paths and given me gifts that I don’t deserve. I thank you Lord for all the blessings you constantly shower on me. Everything is possible with God! I thank Dr. Alan Colman for being my guide and helping me to initiate the work contained in this dissertation. His support and encouragement have been invaluable. Thank you, Alan for your support through the years. Dr. Norris Ray Dunn took me under his wing and guided me through this endeavour. He has been a true mentor, always willing to teach, and I have learned a lot from him. Ray, thank you for showing me the way and helping me reach this juncture. A significant part of the research was conducted at ES Cell International Pte Ltd to whom I would like to express my sincere gratitude. Triona, Jacqui, Robert, Michael, Bruce, Chirag, Suzan and so many others have played important roles and encouraged me at all times. Critical portions of this work were done at the Institute of Medical Biology, A*Star. I would like to register my appreciation for the support and help provided by many people in IMB especially, members of the Ray Dunn lab, Mike Jones lab and Alan Colman lab. Kee Yew, thank you for giving me your precious time and helping me with some of the most important data in this dissertation. -

Supplementary Table S1. PCR Primer and Probe Set Information
Supplementary Table S1. PCR primer and probe set information NCBI Entrez Gene TaqMan Gene Expression Gene Name Gene ID* Symbol Assay ID 59085 Asl argininosuccinate lyase Rn01480437_g1 25698 Ass1 argininosuccinate synthase 1 Rn00565808_g1 292804 Gpi glucose-6-phosphate isomerase Rn01475756_m1 25104 Pc pyruvate carboxylase Rn00562534_m1 361596 Idh2 isocitrate dehydrogenase 2 Rn01478119_m1 81718 Cdo1 cysteine dioxygenasetype 1 Rn00583853_m1 25256 Fmo1 flavin containing monooxygenase 1 Rn00562945_m1 246248 Fmo5 flavin containing monooxygenase 5 Rn00595199_m1 81822 Actb β-actin Rn00667869_m1 *Gene IDs reference The National Center for Biotechnology Information database 1 Supplementary Table S2. Metabolite assignments and H NMR data in urine samples of rat* Urine Metabolite δ1H (ppm) 1 N-Methylnicotinamide 4.48(s) 8.18(t) 8.90(d) 8.97(d) 2 Nicotinamide N-oxide 7.73(dd) 8.12(m) 8.48(m) 3 Formate 8.46(s) 4 Hippurate 3.97(d) 7.55(t) 7.64(m) 7.84(dd) 8.56(s) 5 Tryptophan 3.30(dd) 3.47(dd) 4.05(dd) 7.20(m) 7.28(m) 7.50(d) 7.71(d) 6 Phenylalanine 3.11(dd) 3.28(dd) 3.99(dd) 7.36(m) 7.42(m) 7 Tyrosine 3.04(dd) 3.19(dd) 3.93(dd) 6.89(m) 7.17(m) 8 Fumarate 6.53(s) 9 Allantoin 5.39(s) 6.05(s) 7.26(s) 8.01(s) 10 Urea 5.80(s) 11 Glucose 3.24(dd) 3.39(t) 3.40(t) 3.45(m) 3.48(t) 3.53(dd) 3.70(t) 3.72(dd) 3.76(dd) 3.82(m) 3.84(dd) 3.89(dd) 4.65(d) 5.24(d) 12 Tartrate 4.36(s) 13 Threonine 1.33(d) 3.58(d) 4.24(m) 14 Creatinine 3.05(s) 4.06(s) 15 Creatine 3.02(s) 3.95(s) 16 Glycine 3.57(s) 17 Taurine 3.27(t) 3.43(t) 18 Choline 3.20(s) 3.51(m) 4.06(m) 19 cis-Aconitate -

Fatty Acid Metabolism Driven Mitochondrial Bioenergetics Promotes Advanced Developmental Phenotypes in Human Induced Pluripotent Stem Cell Derived Cardiomyocytes
Fatty acid metabolism driven mitochondrial bioenergetics promotes advanced developmental phenotypes in human induced pluripotent stem cell derived cardiomyocytes Chrishan J.A. Ramachandra1,a,b, Ashish Mehta1,c, Philip Wonga,b,d,e*, K.P. Myu Mai Jaa, Regina Fritsche-Danielsonf, Ratan V. Bhatg, Derek J. Hausenloya,b,h,i,j, Jean-Paul Kovalikb and Winston Shima,b,k* aNational Heart Research Institute Singapore, National Heart Centre Singapore bCardiovascular & Metabolic Disorders Program, Duke-NUS Medical School, Singapore cPSC and Phenotyping Laboratory, Victor Chang Cardiac Research Institute, Sydney, Australia dDepartment of Cardiology, National Heart Centre Singapore eSchool of Materials Science and Engineering, Nanyang Technological University, Singapore fCardiovascular and Metabolic Disease Innovative Medicines and Early Development Unit, AstraZeneca Research and Development, Gothenburg, Sweden gStrategy and External Innovation Department, AstraZeneca, Gothenburg, Sweden hThe Hatter Cardiovascular Institute, University College London, United Kingdom iBarts Heart Centre, St Barthlomew’s Hospital, London, United Kingdom jYong Loo Lin School of Medicine, National University of Singapore kHealth and Social Sciences Cluster, Singapore Institute of Technology 1Both authors contributed equally Running title: Cardiomyocyte metabolism and bioenergetics *Corresponding authors: Philip Wong National Heart Centre Singapore, 5 Hospital Drive, Singapore 169609 Email: [email protected]; Phone: +65 6704 8964; Fax: +65 6844 9053 Winston Shim -

A Novel Approach to Identification of Diagnostic Markers In
A NOVEL APPROACH TO IDENTIFICATION OF DIAGNOSTIC MARKERS IN PROSTATE CANCER by INNA SHYSHYNOVA Submitted in partial fulfillment of the requirements For the degree of Doctor of Philosophy Dissertation Adviser: Professor Andrei Gudkov Department of Biochemistry CASE WESTERN RESERVE UNIVERSITY August, 2006 iii TABLE OF CONTENTS CHAPTER .................................................................................................................. PAGE A NOVEL APPROACH TO IDENTIFICATION OF DIAGNOSTIC MARKERS IN PROSTATE CANCER ...........................................................................................1 TABLE OF CONTENTS.................................................................................................. IV LIST OF FIGURES .......................................................................................................... IX LIST OF TABLES............................................................................................................ XI ACKNOWLEDMENTS .................................................................................................XIII LIST OF ABBREVIATIONS........................................................................................ XIV I. INTRODUCTION ...................................................................................................1 1. Tumor markers.............................................................................................1 2. The prostate..................................................................................................5 -

WO 2016/004043 Al O O©
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2016/004043 Al 7 January 2016 (07.01.2016) P O P C T (51) International Patent Classification: gen Road, Newton, Massachusetts 02459 (US). WOOST- C07K 19/00 (2006.01) A61P 35/00 (2006.01) ER, Richard; 6 Buckskin Lane, Natick, Massachusetts A61K 47/48 (2006.01) 01760 (US). BARDER, Timothy; 14 Waldo Road, Arling ton, Massachusetts 02474 (US). WHALEN, Kerry; 2 1 (21) International Application Number: Hillcrest Road, Waltham, Massachusetts 0245 1 (US). GIF- PCT/US2015/038562 FORD, James; 34 Thorndike St., Somerville, Massachu (22) International Filing Date: setts 02144 (US). 30 June 2015 (30.06.2015) (74) Agents: WARD, Donna T. et al; 142A Main Street, Gro- (25) Filing Language: English ton, Massachusetts 01450 (US). (26) Publication Language: English (81) Designated States (unless otherwise indicated, for every kind of national protection available): AE, AG, AL, AM, (30) Priority Data: AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, 62/019,003 30 June 2014 (30.06.2014) US BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, 62/020,615 3 July 2014 (03.07.2014) US DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, 62/084,306 25 November 2014 (25. 11.2014) u s HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, 62/102,261 12 January 20 15 (12.01.2015) u s KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, (71) Applicant: BLEND THERAPEUTICS, INC.